Light Scattering for the Masses Protein–Protein Interactions
Over-expression of recombinant proteins is commonly used for the production of protein reagents in industry and academia. Problems often occur relating to the stress put on the cells to deal with this huge increase in synthesis. Cellular proteins that are part of the protein synthesis machinery are often up-regulated under such conditions. Large quantities of the recombinant protein can be bound to these cellular proteins, making purification difficult.
Over-expression of recombinant proteins is commonly used for the production of protein reagents in industry and academia. Problems often occur relating to the stress put on the cells to deal with this huge increase in synthesis. Cellular proteins that are part of the protein synthesis machinery are often up-regulated under such conditions. Large quantities of the recombinant protein can be bound to these cellular proteins, making purification difficult.
Scale-up purification of a 61 kDa recombinant protein was found to contain a significant contaminant (Figure 1), not found in the smaller scale trials. The 28 kDa protein was identified by Edman degradation as rotamase, an enzyme that assists in protein folding. Co-purification of the rotamase suggests it is bound to the protein of interest, thus preventing further purification with techniques such as size exclusion chromatography (SEC).
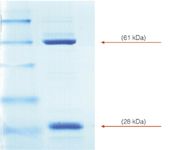
Figure 1: SDS-PAGE of a partially purified recombinant protein (Mw = 61 kDa) containing a significant contaminant of the enzyme rotamase (Mw = 28 kDa).
Analytical size exclusion chromatography, followed by on-line direct mass detection by multiangle light scattering (MALS), showed that a significant amount of the recombinant protein was not bound to the rotamase. The sample was separated with a YMC-pack Diol-200 column and detected by Wyatt Technology's 18-angle DAWN photometer and the Optilab-DSP Interferometric
Refractometer.
Three peaks were isolated that contained 93 kDa, 68 kDa and 28 kDa species (Figure 2). The DAWN was able to identify the peaks in the following way: a non-covalent complex of one rotamase molecule and one recombinant protein molecule (peak 1), the free recombinant protein (peak 2) and the free rotamase (peak 3).
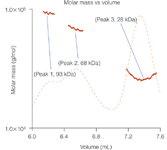
Figure 2: Peak 1, with a molar mass of 93 kDa is one recombinant protein and one rotamase; Peak 2, with a molar mass of 68 kDa, is a free recombinant protein; Peak 3 is a 27 kDa free rotamase.
Estimating the molar masses of the proteins using columns calibrated to a set of protein standards (BioRad) resulted in significantly different values for the three peaks: 146 kDa, 98 kDa, and 46 kDa, respectively. Thus, the information provided by MALS system — an absolute system because it is not calibrated with standards — contributed to the correct interpretation of the chromatogram, provided invaluable insight for development of the scale-up purification protocol and into the protein–protein interactions.
We are most grateful to Nestor B. Nestor and George A. Karam, Pfizer Central Research, Groton, Connecticut, USA, for their efforts in preparing this application note. Protein–Protein Interactions.

Wyatt Technology Corporation
6300 Hollister Avenue, Santa Barbara, California 93117, USA
tel. +1 805 681 9009 fax +1 805 681 0123
E-mail: info@wyatt.com
Website: www.wyatt.com

Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.
Extracting Estrogenic Hormones Using Rotating Disk and Modified Clays
April 14th 2025University of Caldas and University of Chile researchers extracted estrogenic hormones from wastewater samples using rotating disk sorption extraction. After extraction, the concentrated analytes were measured using liquid chromatography coupled with photodiode array detection (HPLC-PDA).



















