Separation Science: The State of the Art: New Frontiers in Lipidomics
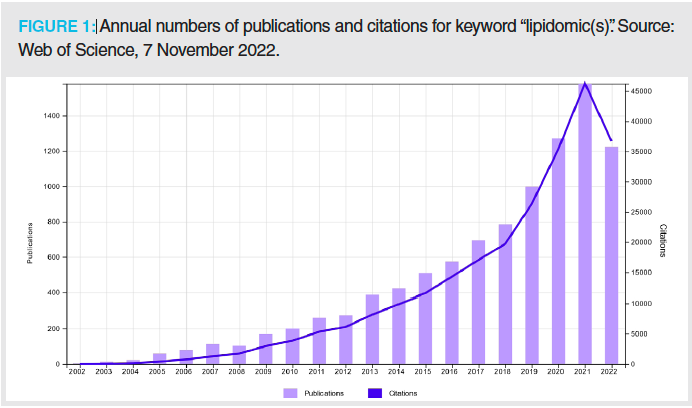
The first use of the term “lipidomic(s)” was in 2002, and up until now, almost 10,000 papers on lipidomics have been published (Figure 1), which clearly illustrates that lipidomics is probably the fastest growing omics discipline at present. However, this fast growth is also accompanied by some negative issues. Many newcomers and beginners are entering the lipidomic field, often without adequate knowledge of basic analytical methodology, nomenclature, and data reporting, which can cause confusion in data interpretation and reporting (1). Recently, the International Lipidomics Society (ILS) has been established with the goal of harmonizing the field, unifying the nomenclature by updating shorthand annotations in cooperation with the LIPID MAPS consortium (2), and introducing a checklist system for transparent reporting of lipidomic data (3). The first Gordon Research Conference on Lipidomics was organized in August 2022 by John Bowden and Kim Ekroos. A smart example of the recognition of lipidomic research is the fact that three distinguished lipidomic researchers (Erin Baker, Shane Ellis, and Livia Eberlin) were awarded prestigious medals at the last IMSC 2022 conference in Maastricht, Netherlands. This is just a brief illustration—and certainly not complete—that the lipidomic community has started to be active in the organization of various events and activities to promote further growth of the field.
The driving force of any research field must be scientific progress in the methodology, which may bring new applications. The lipidomic community pays considerable attention to proper quantitation using appropriate exogenous internal standards (4), including the validation of analytical methods and the use of quality controls (5). The broad consensus is the use of at least one internal standard for each lipid class to be quantified in case of lipid class separation (hydrophilic interaction liquid chromatography [HILIC] and ultrahigh-performance supercritical fluid chromatography [UHPSFC]) or shotgun approaches, but the lipid species separation techniques (reversed-phase ultrahigh-pressure liquid chromatography [UHPLC]) require more. The second important aspect is the lipidomic coverage. Biologists estimate the presence of about 100,000 lipid species in Nature, but the best analytical methods typically report less than 1000, so there is considerable room for improvement in the analytical methodology. However, one method cannot cover this complexity; therefore, we need several complementary analytical methods and sample preparation protocols. Lipidomic analysis is strongly dependent on mass spectrometry (MS) and chromatographic techniques. The main advantage of chromatography is the separation of various isomeric and isobaric lipids, which is sometimes easier than with MS-based approaches. The prevailing technique is UHPLC, but more groups realize the potential of UHPSFC (6), which is perfectly suited for molecules with large hydrophobic parts, such as lipids. From an MS point of view, many researchers appreciate the potential of ultrahigh‑resolving power mass spectrometers, typically orbital trap analyzers, and MS imaging for the visualization of tissues. The increasing resolving power in ion mobility has shown its full potential, which may be easily coupled with liquid separation techniques and MS (7). One trend that cannot be overlooked in lipidomic analysis is the shift towards more structural details, such as data on the fatty acyl level or even the double bond level, which could reveal new biological information previously unknown (8).
Multiomics integration of lipidomics with other omics approaches, such as metabolomics, proteomics, transcriptomics, and genomics, is the research direction of the future. Concentrations of lipid species are influenced by the activity of the corresponding enzymes responsible for the particular metabolic pathways involved in the biosynthesis and subsequent degradation of lipids. RNA—both coding and noncoding—shows a correlation with lipidome. The typical example is an alteration of miRNA in cancer. Known genetic mutations have connections to serious human diseases, which is also reflected in the dysregulation of particular metabolic pathways. The integration of multiple omics data is needed for the full understanding of the biological mechanisms of the diseases studied, but such research requires experts for each omics technique and a strong bioinformatic background, which is still in its infancy. I believe that the substantial effort invested in this area will result in fast progress and a better understanding of the biological roles of lipids in disease and health, with possible consequences in biomarker discovery and drug development.
The driving force for progress in lipidomic analysis is biomedical applications and clinical analyses because the translation of key discoveries from academic research laboratories into the real clinical environment could bring substantial benefits to clinicians. Many research groups around the world are working hard to investigate the potential of lipidomics in biomarker discovery applicable to early disease detection, the monitoring of therapeutic response, and helping to select the best possible therapy for a given phenotype, which is known as personalized medicine. The expectations are enormous, but unfortunately, successful clinical translation is rather rare. First, we must harmonize the molar concentrations reported by individual laboratories with their individual workflows, which is the main goal of a running ring trial on human plasma lipidome organized by the Clinical lipidomics Interest Group (CLIG) within ILS. I know of only one example of completed translation from the research laboratory into real clinical use, which is the ceramide test for cardiovascular risk (9) implemented by the Mayo Clinic. At present, the spin-off company created by my university and the FONS company is working on the clinical validation of our lipidomic test for early detection of pancreatic cancer based on blood analysis (10). The methodology is ready to use, with a sample throughput of 20,000 samples/year, but it will still take several years to collect a sufficient number of samples from high-risk subjects to prove the real benefit for the clinical management of cancer patients. It is interesting to note that we observe similar dysregulation patterns for other types of cancer (11). However, the biological mechanism of the observed lipidomic dysregulation has not yet been discovered and will require suitable biological models and a multiomics approach. Another exciting discovery was published by the Fendt group (12) that demonstrated that cancer cells exhibit enormous flexibility, which may result in the desaturation of fatty acyls in unusual positions not common for normal cells. I list here only examples of lipidomic research for cancer and cardiovascular diseases, but there are many examples in the literature about changed metabolic pathways for various lipids in other diseases, such as liver diseases and brain disorders.
References
1) H.C. Köfeler et al., Nat. Com. 12, 4771 (2021).
2) G. Liebisch et al., J. Lipid Res. 61, 1539 (2020).
3) J.G. McDonald et al., Nat. Met. 4, 1086 (2022).
4) M. Holčapek, G. Liebisch, and K. Ekroos, Anal. Chem. 90, 4249 (2018).
5) D. Wolrab et al., Anal. Bioanal. Chem. 412, 2375 (2020).
6) D. Wolrab et al., Trends Anal. Chem. 149, 116546 (2022).
7) K.L. Leaptrot et al., Nat. Com. 10, 985 (2019).
8) R.S.E. Young et al., Frontiers Endocrin. 12, 689600 (2021).
9) R. Laaksonen et al., Eur. Heart J. 37, 1967 (2016).
10) D. Wolrab et al., Nat. Com. 13, 124 (2022).
11) D. Wolrab et al., Sci. Rep. 11, 20322 (2021).
12) K. Vriens et al. Nature 566, 403 (2019).
Michal Holčapek is a professor at the University of Pardubice, Faculty of Chemical Technology, Department of Analytical Chemistry, in Pardubice, Czech Republic.

New Study Investigates Optimizing Extra-Column Band Broadening in Micro-flow Capillary LC
March 12th 2025Shimadzu Corporation and Vrije Universiteit Brussel researchers recently investigated how extra-column band broadening (ECBB) can be optimized in micro-flow capillary liquid chromatography.