Separation Science: The State of the Art: Graph Neural Networks for Improved Retention Time Predictions
To speed up the optimization of chromatographic separations—a process often referred to as method development—models that aim to predict the retention time or retention factor of a compound are frequently used. These models can be empirical or mechanistic in nature, and predict the retention of a compound based on one or more chromatographic parameters, such as the fraction of organic modifier in the mobile phase in isocratic separations, the gradient profile in gradient separations, or the pH or temperature of the mobile phase (1). Such models are typically established based on a number of scouting runs or exploratory runs, wherein the parameter under investigation, for example, the fraction
of organic modifier in the mobile phase, is varied a number of times—a number that is typically equal to or higher than the number of parameters in the retention model. Thereafter, a retention model is built by relating the experimentally obtained retention factors to the chromatographic parameter. Well-known examples of such retention models are the linear solvent strength model (2) and the Neue-Kuss model (3). An advantage of this approach is that (structural) information about the compound under consideration is not required. A disadvantage is that multiple informative scouting runs for each compound need to be executed before the model can be built and used, which takes time.
Alternatively, when structural information about the compound under consideration is available, quantitative structure retention relations (QSSR) can be used for retention time or retention factor predictions (4). QSSR are mathematical relations between the retention time of a compound and its structural features. Typically, QSSR are built for a specific chromatographic setup, that is, a fixed combination of stationary phase and mobile phase conditions for a large number of compounds with known structures. The obtained retention times are then correlated with the compounds’ structural features or descriptors to build a retention model. Once the model is available, the retention times of new compounds can be predicted without having to run any new experiments, significantly decreasing the time required to optimize separations. However, before the model becomes available, the retention times of typically at least 50–100 compounds on the particular chromatographic setup are required, while another prerequisite is that the structure of the compound should be known.
To build adequate QSRR, structural features or descriptors that adequately represent the interactions between the compounds under consideration and the chromatographic conditions need to be selected. These descriptors can be physicochemical, quantum mechanical, or topological in nature. Examples include molecular mass, carbon number, polarizability, and (calculated) partition coefficient, although thousands of possible candidates exist (5). Once suitable features have been selected, they are mapped against the experimental retention times or retention factors using regression models or machine learning algorithms, such as multi-layer perceptrons (MLP), random forests (RF), or support vector machines (SVM), to obtain a retention model (6).
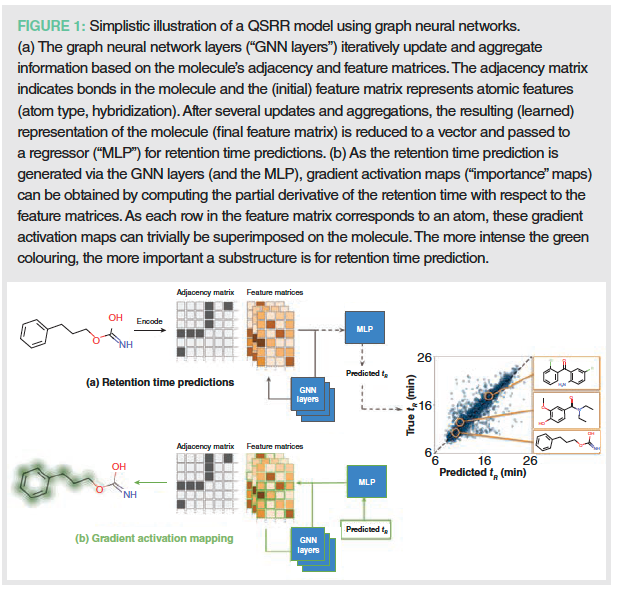
More recent additions to these approaches are graph neural networks (GNNs). As opposed to the more traditional machine learning algorithms, GNNs use information about the atoms themselves and their bonds to adjacent atoms, to obtain optimized and meaningful molecular representations for retention time predictions. This is done by first encoding the atoms numerically. These atom encodings subsequently undergo a transformation and aggregation step to incorporate information about their neighbouring atoms via a set of learnable weight matrices. This is repeated a number of times (layers) for increasing distances (radii). This allows the GNN to learn complex relations between atoms or molecular sub-structures over both short and longer distances, leading to powerful and meaningful molecular representations. The obtained atom encodings are finally reduced into one-dimensional vector representations of the molecules, which are then inputted to a regression model (such as MLP) to produce retention time predictions. A simplified illustration of a GNN is shown in Figure 1(a).
We recently demonstrated the potential of GNNs for retention time predictions in different separation modes (reversed-phase and hydrophilic interaction liquid chromatography) (7). When compared with more traditional approaches, such as MLP, RF, and SVM, GNNs generally performed better than the traditional approaches, with mean absolute errors (MAEs) between the experimental and predicted retention times that were typically 5–25% lower than those obtained with the traditional models, depending on the dataset. This was attributed to the fact that GNNs result in a more expressive and discriminative molecular representation for retention time predictions, due to the learnable molecular representation based on the large number of atom and bond features. It was, however, also observed that GNNs in some cases suffer from some unusually high prediction errors. This was attributed to a possible overfitting of the GNNs to (some of) the training data, which could be explained by the large number of weights that need to be trained at each layer of the GNN. In future research, this could be tackled by adjusting the number of learnable weights or increasing the size of the datasets.
To better understand what molecular substructures contribute to the retention time predictions, so-called gradient activation maps (GAMs) can be computed from the GNNs (8). Visualizing these GAMs can help understand to what part of the molecule the model is “looking” when it makes its retention time prediction and as such help understand what substructures contribute to retention under particular chromatographic conditions. For an example, see Figure 1(b).
In conclusion, GNNs show great promise as new machine learning tools for retention time predictions using QSRR and certainly deserve further exploration. Researchers who are interested in creating their own GNNs for retention time predictions of small molecules are welcomed to use the recently developed open source Python package MolGraph, which can be installed from: https://github.com/akensert/molgraph
References
1) M.C. García-Alvarez-Coque, G. Ramis-Ramos, J.R. Torres-Lapasió, and C. Ortiz-Bolsico, Analytical Separation Science 1, 199–226 (2015).
2) L.R. Snyder, J.W. Dolan, and J.R. Gant, J. Chromatogr. A 165(1), 3–30 (1979).
3) U.D. Neue and H.-J. Kuss, J. Chromatogr. A 1217(24), 3794–3803 (2010).
4) R.I.J. Amos, P.R. Haddad, R. Szucs, J.W. Dolan, and C.A. Pohl, TrAC Trends Anal. Chem. 105, 352–359 (2018).
5) R. Kaliszan, Chem. Rev. 107(7), 3212–3246 (2007).
6) R. Bouwmeester, L. Martens, and S. Degroeve, Anal. Chem. 91(5), 3694–3703 (2019).
7) A. Kensert, R. Bouwmeester, K. Efthymiadis, P. Van Broeck, G. Desmet, and D. Cabooter, Anal. Chem. 93(47), 15633–15641 (2021).
8) P.E. Pope, S. Kolouri, M. Rostami, C.E. Martin, and H. Hoffmann, 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 10764–10773 (2019).
Alexander Kensert is a post-doc in the Department of Pharmaceutical and Pharmacological Sciences, University of Leuven (KU Leuven), Belgium.
Gert Desmet is a full professor in the Department of Chemical Engineering, Vrije Universiteit Brussel, Belgium.
Deirdre Cabooter is a professor in the Department of Pharmaceutical and Pharmacological Sciences, University of Leuven.

How Many Repetitions Do I Need? Caught Between Sound Statistics and Chromatographic Practice
April 7th 2025In chromatographic analysis, the number of repeated measurements is often limited due to time, cost, and sample availability constraints. It is therefore not uncommon for chromatographers to do a single measurement.
Fundamentals of Benchtop GC–MS Data Analysis and Terminology
April 5th 2025In this installment, we will review the fundamental terminology and data analysis principles in benchtop GC–MS. We will compare the three modes of analysis—full scan, extracted ion chromatograms, and selected ion monitoring—and see how each is used for quantitative and quantitative analysis.
Rethinking Chromatography Workflows with AI and Machine Learning
April 1st 2025Interest in applying artificial intelligence (AI) and machine learning (ML) to chromatography is greater than ever. In this article, we discuss data-related barriers to accomplishing this goal and how rethinking chromatography data systems can overcome them.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)