Liquid Chromatography Methods for the Separation of Short RNA Oligonucleotides
LCGC Europe
Synthetic oligonucleotides have become increasingly popular as a result of the recent discovery of ribonucleic acid interference (RNAi), a natural process for silencing gene expression.
Pages 632-639
Synthetic oligonucleotides have become increasingly popular as a result of the recent discovery of ribonucleic acid interference (RNAi), a natural process for silencing gene expression. As biomedical researchers evaluate the use of antisense and small interfering RNAs (siRNAs) as potential therapies for the treatment of disease, the need for improved methods for the chromatographic separation and analysis of oligonucleotides has become apparent. This article presents a review of different liquid chromatography (LC) methods and strategies for the chromatographic separation of short RNA oligonucleotides.
There has been considerable interest recently in the use of synthetic oligonucleotides as potential therapeutic agents capable of suppressing the synthesis of specific proteins (1–3). Targeted "knockdown" of specific gene products using an antisense ribonucleic acid (RNA) strategy dates back to the late 1990s (4). In this approach, a single-stranded oligonucleotide complementary to the messenger RNA (mRNA) encoding a targeted protein leads to disruption of ribosomal transcription and protein synthesis. In theory, antisense oligonucleotides can be applied to any disease in which protein overexpression is detrimental, and a number of antisense oligonucleotides have been evaluated as potential therapies (5). The need for long complementary oligonucleotides and the stoichiometric nature of mRNA inactivation (one antisense molecule: one mRNA inactivation) places considerable constraints on developing cost-effective antisense drugs.
The more recently discovered small interfering RNA (siRNA) mechanism for silencing gene expression involves a short double-stranded RNA molecule of about 21 base pair length, which activates the RNA interference (RNAi) silencing pathway (6,7), thereby achieving catalytic degradation of the target mRNA (one siRNA molecule inactivates multiple mRNAs). An overview of the RNAi pathway for targeted gene silencing is illustrated in Figure 1.
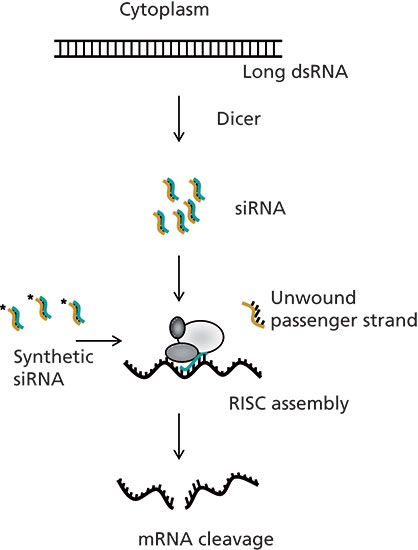
Figure 1: RNA interference mechanism. Long dsRNA in the cytoplasm is cleaved into 21-mer strands (siRNA) by the protein, Dicer. Small interfering RNA is incorporated into the RNA-induced silencing complex (RISC), where the passenger strand is unwound and degraded leaving the guide strand bound to RISC. The RISC-guide strand complex base-pairs with a complementary sequence of the mRNA and induces cleavage of the mRNA, thereby preventing protein translation. Synthetic siRNAs can be introduced into the cell and achieve the same action in the RNAi mechanism.
The discovery of siRNA gene silencing in animals (8) and human cells (9) has led to a surge of interest in the use of siRNA for biomedical and drug development research. Many biomedical and pharmaceutical companies have become involved in the exploration of the preparation and use of siRNAs as potential therapies for the treatment of diseases such as cancer, macular degeneration, and viral infections (10).
Oligonucleotide and siRNA Structure and Preparation
RNA is a biologically important molecule that consists of a long chain of nucleotide units. Each nucleotide contains a ribose sugar, a nitrogenous base, and a phosphate group. There are four bases in RNA: adenine (A), guanine (G), cytosine (C), and uracil (U) (Figure 2). Oligonucleotides are short, single-stranded RNA or deoxyribonucleic acid (DNA) molecules that can readily bind, in a sequence-specific manner, to their respective complementary oligonucleotides to form duplexes. Small interfering RNA is a small double-stranded RNA (usually 21 nucleotides) with two nucleotide overhangs on each 3'-end. Each strand has a 5'-phosphate group and a 3'-hydroxyl group (Figure 3).
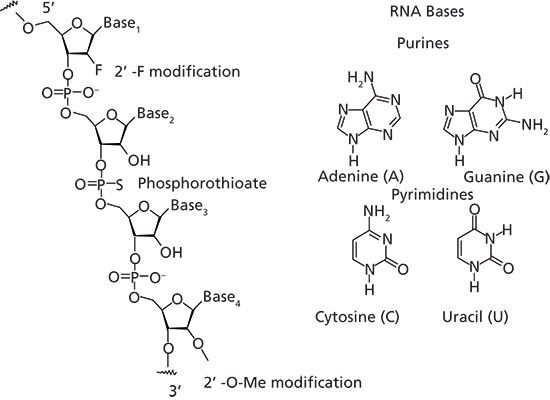
Figure 2: Oligonucleotide structure. Different modifications include phosphothioate backbone modification where one oxygen atom on the phosphodiester backbone is replaced with a sulphur atom, and 2'-sugar modifications, such as 2'-F and 2'-O-Me. The four bases in RNA are adenine (A), guanine (G), cytosine (C), and uracil (U).
Various chemical modifications are often made to synthetic oligonucleotides to prevent attack by nucleases, which can lead to siRNA degradation and instability (11,12). Incorporation of either a fluoro or methoxy group into the 2' position of the sugar or the use of a phosphothioate linkage is commonly used to improve siRNA stability (Figure 2) (13). In the phosphothioate modification, oxygen in the phosphodiester linkage is replaced with a sulphur atom. This introduces an additional stereocenter into the molecule giving rise to two possible diastereomers for every phosphothioate linkage, and making the resulting oligonucleotide sample mixtures highly complex and very difficult to chromatographically resolve. All of these modifications help to improve oligonucleotide stability while retaining, and sometimes even increasing, their silencing activity. These modifications also tend to increase the hydrophobicity of the oligonucleotides, while also increasing the temperature at which the duplex melts (Tm) into its corresponding single strands.
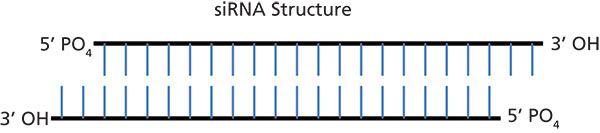
Figure 3: Small interfering RNA (siRNA) structure. A 21-mer siRNA is shown with two nucleotide overhangs on each 3'-end. Each strand has a 5'-phosphate group and a 3'-hydroxyl group. The siRNA duplex consists of two complementary strands, the sense (or passenger) and antisense (or guide) strands. In RNA, adenine base-pairs with uracil by forming two intermolecular hydrogen bonds (A–U) and guanine base-pairs with cytosine by forming three intermolecular hydrogen bonds (G–C).
Oligonucleotides are readily synthesized via stepwise synthesis using phosphoramidite chemistry with automated solid-phase synthesizers (14). Although the individual synthetic process reactions can be very efficient and provide high yields, the total number of synthetic steps for making a 21-mer RNA can be more than 80 chemical steps (with about four chemical steps for each cycle). Consequently, because of the accumulation of many small errors, the final oligonucleotide product typically contains a variety of closely related impurities that can be very difficult to separate and remove during final product purification (15). Some of the most common impurities include sequence deletions, such as n–1, n–2, and so on, where one or more nucleotide fails to attach to the sequence during synthesis. In addition, depurination, oxidation, and other chemical modification or degradation of the nucleotide bases can lead to a variety of closely related impurities that can be very challenging to resolve from the desired product. When dealing with double-stranded siRNAs, the sample mixtures can become even more complex with each strand introducing its own set of impurities. These impurities include mismatched sequences and noncomplementary single stranded sequences. The presence of these impurities in a therapeutic mixture can lead to unwanted, nontargeted gene silencing, while the presence of any nonhybridized single-stranded RNAs can also lead to a decrease in therapeutic potency (40). Therefore, when developing siRNA therapeutics, one of the major challenges is ensuring good purity to minimize off-target gene silencing effects. Consequently, developing good chromatographic techniques is often critically important in the oligonucleotide drug development process.
Chromatographic Separation of Oligonucleotides
When developing analytical methods for the separation of oligonucleotides, some of the unique features of these molecules need to be considered. First, the pKa of the phosphodiester linkage is 2, meaning that in aqueous solution above pH 4 these molecules contain one negative charge for every phosphodiester linkage - that is, a 21-mer oligonucleotide contains 21 negative charges at pH 7. Therefore, traditional reversed-phase liquid chromatography (LC) conditions tend not to work well, and ion-pair reversed-phase LC or anion-exchange chromatography techniques need to be considered. Secondly, many single-stranded RNA oligonucleotides can form higher order (tertiary) structures including bends, loops, dimers, or other aggregates, and elevated chromatographic temperatures must be used to "melt" such structures, allowing for efficient analysis (16). Also, when analyzing single-strands versus duplex samples, different methods must be considered, with single-strand analysis typically taking place at temperatures >60 °C and duplex samples typically being analyzed at or below room temperature. A typical siRNA duplex LC analysis is shown in Figure 4. Finally, since RNA and DNA molecules both have a strong absorbance at 260 nm, these samples are often analyzed using UV detection, although fluorescence and especially mass spectrometry (MS) detection (17) are also quite important for RNA analysis.
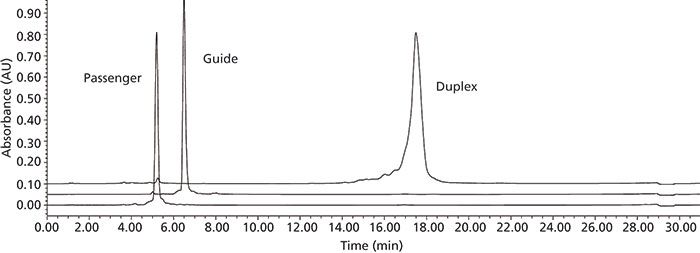
Figure 4: A typical UHPLC analysis of duplex 21-mer siRNA. Column: 100 mm × 2.1 mm, 1.7-μm dp Waters CSH C18; mobile-phase A: 400 mM HFIP–16.3 mM TEA in water (pH 7.9); mobile-phase B: methanol; segmented linear gradient: 25–33% B in 10 min, 33–36% B in 20 min, 36–60% B in 28 min, 3-min equilibration at 25% B; total run time: 31 min; flow rate: 0.3 mL/min; detection: UV absorbance at 260 nm; column temperature: 15 °C.
A number of chromatographic methods have been reported for the analysis and purification of oligonucleotides, including capillary gel electrophoresis (CGE) (18,19), anion-exchange high performance liquid chromatography (HPLC) (20–33), ion-pair reversed-phase LC (34–51), and mixed-mode LC (52–56). These different LC techniques are reviewed in the following sections.
Ion-Exchange Liquid Chromatography: Ion-exchange chromatography is an excellent method for separating charged molecules, and is a commonly used chromatographic method for the separation of multiply charged oligonucleotides (20). In ion-exchange chromatography, separation is based on the differential electrostatic affinities of charged molecules for a charged stationary phase. For the separation of highly negatively charged oligonucleotides, anion-exchange chromatography is used with positively charged stationary phases that exchange the negatively charged oligonucleotides through competition with anions from the mobile phase. As a result, longer oligonucleotides with greater charge are more strongly retained on the column and shorter oligonucleotides, which have progressively fewer negative charges, have progressively shorter retention. This method is especially useful for the separation of the very common N–x deletions of oligonucleotides (Figure 5[a]), but is less useful for detecting subtle changes on the full-length sequence that do not alter the total number of charges (Figure 5[b]).
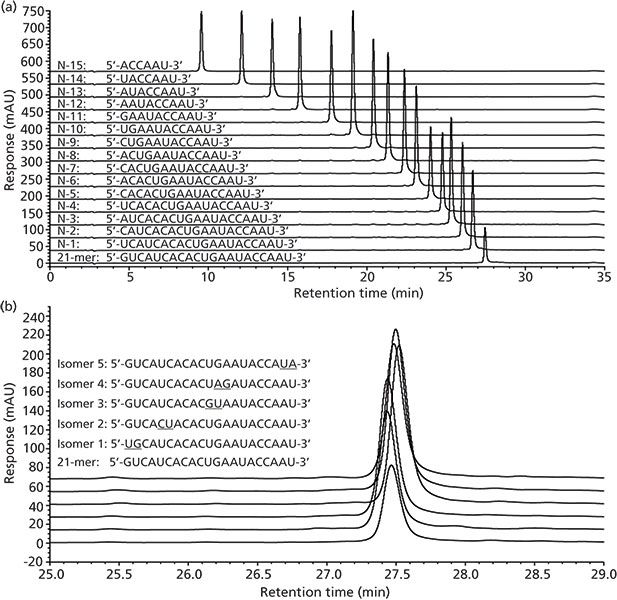
Figure 5: Chromatographic separation of oligonucleotide standards by strong-anion-exchange LC: (a) Separation of N–x deletion series, (b) separation of "base-flip" isomer standards (25–29 min portion of chromatogram shown). Column: 150 mm × 4.6 mm, 3-μm dp Proteomix SAX-NP3 (Sepax Technologies); mobile-phase A: 80:20 (v/v) 10 mM Tris (pH 8)-ethanol; mobile-phase B: 600 mM sodium bromide salt concentration in mobile-phase A; linear gradient: 20–80% B over 30 min with 5-min column equilibration at 20% B; flow rate: 0.5 mL/min; column temperature: 60 °C; injection volume: 15 μL; detection: UV absorbance at 260 nm. Adapted with permission from reference 59.
Successful separation of small oligonucleotides and their sequential analysis by anion-exchange HPLC was originally reported more than 30 years ago (21,22). In these studies, anion-exchange columns, such as Permaphase AEX (DuPont) or Partisil SAX (HiChrom), with a salt gradient using phosphate or acetate buffers were used. Later, Pingoud and colleagues (23) showed the use of strong-anion-exchange LC for the excellent resolution of longer oligonucleotides (up to 64 bases) from their N–1 deletion products, using a Whatman SAX column and a phosphate salt gradient. Over the years, improved types of anion-exchange resins were developed to improve the resolution of oligonucleotides with strong-anion-exchange LC. Alkylamine derivatized (24) and polyethyleneimine (PEI) coated (25,26) porous silica phases were prepared for the anion-exchange HPLC separation of oligonucleotides, where resolution of oligonucleotides of up to 30 bases from their N–1 deletion products was obtained. Optimization of the PEI bonding chemistry with quaternization of the ion-exchange matrix was shown to further increase the resolution of N–1 deletion products for up to 50-mer oligonucleotides (27).
For chromatographic separations using harsher conditions such as elevated column temperatures and neutral-to-high pH mobile phases, the relative chemical instability of silica-based stationary phases can be a significant disadvantage, because particle erosion and column degradation can lead to a significant loss of performance over time (28). Consequently, PEI-coated porous zirconia stationary phases were provided by Professor Carr for use as anion exchangers in the strong-anion-exchange LC of oligonucleotides (29). These stationary phases were able to provide single-nucleotide unit resolution for oligonucleotides of up to 50-nucleotide length. Furthermore, the zirconia-based stationary phases can be operated at elevated column temperatures of 75 °C, which allow for the highly advantageous elution of oligonucleotides with a low ionic strength mobile phase; those conditions lead to loss of performance and bed collapse with conventional silica-based columns.
The development of nonporous anion exchangers for use in strong-anion-exchange LC of proteins (30) and oligonucleotides (31) is another important milestone. These nonporous columns, such as TSKgel (Tosoh Biosciences), were prepared by introducing diethylaminoethyl (DEAE) groups into nonporous spherical hydrophilic resins with 2.5-μm diameter particles. The effects of some of the critical chromatographic parameters in strong-anion-exchange LC for oligonucleotides were also studied. Examination of the eluent pH showed that pH should ideally be ≥8.5 for the separation of oligonucleotides. Furthermore, the range of pH 8.5–9.5 was shown to be better suited for separations based on differences in chain length, while a pH of 10.5 was better suited for separation based on differences in base composition. Because these stationary phases are chemically very stable, operating at high pH would not cause any problems. The addition of salts, such as sodium chloride or sodium perchlorate, also has an influence on chromatographic separation. Finally, other useful methacrylate-based (polymeric) anion-exchange stationary phases have also been developed and successfully used for oligonucleotide separations (32–34).
Ion-Pair Reversed-Phase Liquid Chromatography: Ion-pair reversed-phase LC is another very commonly used LC technique for the analysis and separation of oligonucleotides (35). In ion-pair reversed-phase LC, negatively charged oligonucleotides interact with positively charged alkylammonium ions in a way that permits the chromatographic separation of the former on a reversed-phase stationary phase. Different mechanisms regarding the ion pair interaction have been proposed, depending on whether the ion-pair process occurs in the mobile phase (36) or in the stationary phase (37). In the first proposed mechanism, ion-pair formation occurs in the aqueous mobile phase, and the neutralized ion pair is then adsorbed onto the hydrophobic stationary phase, with retention controlled by the overall hydrophobicity of the ion pair. In the second mechanism, the unpaired ion (such as an alkylammonium ion) from the mobile phase is adsorbed onto the stationary phase, which then acts as an ion-exchange stationary phase for the separation of charged oligonucleotides.
In ion-pair reversed-phase LC, in addition to the charge–charge interactions, hydrophobic interactions from the individual bases also significantly contribute to the overall oligonucleotide retention. The hydrophobicity of oligonucleotide bases follows the order C < G < A < T (for DNA-based oligonucleotides), with cytosine being the least hydrophobic base (38,39). Therefore, in addition to the oligonucleotide length, the retention of oligonucleotides also depends on the specific base composition, where the overall hydrophobicity is the sum of all the bases in the sequence. As such, ion-pair reversed-phase LC can be especially useful for separating impurities from changes in the full-length sequence such as depurinations and other chemical modifications on the bases. Separation of an oligonucleotide and a series of its deletions (N–1 to N–15) are shown in Figure 6(a). This illustrates the retention of oligonucleotides by ion-pair reversed-phase LC in which the longer oligonucleotides are more retained, but overall retention is mostly governed by the base composition, where cytosine (the least hydrophobic) base deletions resulted in increased retention. Additionally, the ion-pair reversed-phase LC method provided some separation of very subtle differences on the oligonucleotide sequence, such as the reversal of two neighbouring bases at different locations along the sequence (Figure 6[b]).
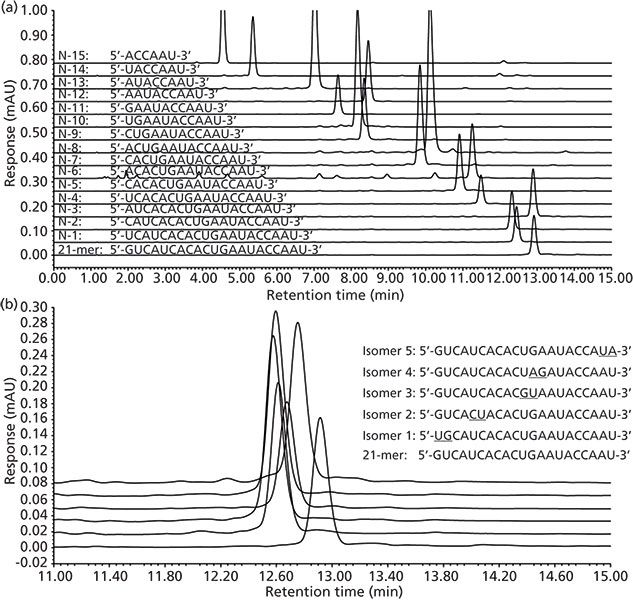
Figure 6: Chromatographic separation of oligonucleotide standards by ion-pair reversed-phase LC: (a) Separation of N–x deletion series, (b) separation of "base-flip" isomer standards (11–15 min portion of chromatogram shown). Column: 150 mm × 4.6 mm, 3.5-μm dp XBridge C18 (Waters); mobile-phase A: 100 mM TEAA in water; mobile-phase B: 100 mM TEAA in acetonitrile; linear gradient: 5–10% mobile phase B over 15 min with 5-min column equilibration at 5%; flow rate: 1.5 mL/min; column temperature: 65 °C; injection volume: 15 μL; detection: UV absorbance at 260 nm. Adapted with permission from reference 59.
Triethylammonium acetate (TEAA) is one of the most commonly used ion-pair reagents for the separation of oligonucleotides because of its good separation efficiency (40,41). Typical TEAA concentrations in the aqueous solution are 100 mM at pH 7 with acetonitrile as the organic solvent. Gilar and colleagues (42) did an extensive study of the prediction of retention for oligonucleotides with ion-pair reversed-phase LC using TEAA at pH 7 as the ion-pairing reagent. Using 39 different oligonucleotides, they demonstrated the successful application of a model for the prediction of the mobile phase strength required to elute the oligonucleotides. Shallow linear gradients of organic modifiers are typically used for these separations because studies have shown that there can be a sharp change in the retention factor (k) of oligonucleotides with a small change in the mobile phase strength. Gilar and colleagues (42) reported that the retention factor for a 15-mer oligonucleotide decreased from 100 to 13.5 to 3.2 with a very small change of mobile-phase composition from 8% to 9% to 10% acetonitrile.
Another very useful ion-pairing mobile phase is the combined hexafluoroisopropanol (HFIP) and triethylamine (TEA) buffer system proposed by Apffel and colleagues (43). This HFIP-based separation uses methanol as the organic modifier because HFIP is immiscible with acetonitrile and miscible with water, methanol, isopropanol, and hexane. Using 400 mM HFIP and adjusting the solution to pH 7.0 with TEA, separations comparable to those obtained with the 100 mM TEAA mobile phases were also obtained. The main difference, however, was in the electrospray ionization (ESI) performance for the MS detection of oligonucleotides. Comparison of different solvent systems, such as 400 mM HFIP adjusted to pH 7.0 with TEA, water, 100 mM TEAA (pH 7), and 25 mM TEA (pH 10), showed superior MS signal with the 400 mM HFIP buffer system, compared to significantly suppressed MS signal with the 100 mM TEAA buffer. This significant difference in MS detection was attributed to the dynamic adjustment of the pH in the ESI droplet as a function of the removal of the anionic counterion from the droplet by evaporation. First, comparing the volatilities for the two different buffer systems, HFIP (boiling point [bp] = 57 °C) is more volatile than TEA (bp = 89 °C), with acetic acid being the least volatile (bp = 118 °C). Secondly, the weak acid and base system with HFIP and TEA maintains a stable pH at ~7.0. The pKa values of acetic acid, HFIP, and TEA are 4.75, ~9, and 11.01. Therefore, at pH 7.0, acetic acid is completely dissociated and it cannot be removed by evaporation on the MS source, whereas HFIP is not charged and it can be evaporated freely. Furthermore, the mechanism proposed by Apffel suggests that during the separation, the TEA ions ion-pair with the negatively charged phosphates on the oligonucleotide backbone, because the more volatile HFIP is evaporated from the droplet surface causing the pH on the surface to rise (~ pH 10). As the pH rises, the oligonucleotide–TEA ion pair dissociates, and the oligonucleotide can be desorbed into the gas phase. In addition, the role of TEA is very important in this mechanism because, in general, oligonucleotides have a high binding affinity for Na+ and K+ cations on the polyanionic phosphate backbone and these cation adducts can diminish the sensitivity for electrospray ionization. The use of a strong base such as TEA effectively suppresses the sodium and potassium adducts formation by a displacement mechanism and consequently dramatically increases the ESI sensitivity. This HFIP and TEA mobile-phase system is now routinely used for oligonucleotide analysis with ion-pair reversed-phase LC and ESI–MS detection (43–50).
Gilar and colleagues (47) further evaluated the HFIP and TEA buffer system and showed that concentration of the ion-pairing TEA ion, rather than the concentration of HFIP, in the mobile phase plays a critical role in the separation. They extensively studied the effect of the concentration of both TEA and HFIP in the mobile phase by varying the TEA concentration from 0.56 to 31.4 mM range (pH 7–9), and the HFIP concentration from 12.5 to 400 mM, using oligonucleotides up to 30-mer length. They showed that the HFIP and TEA buffer system was most effective at 400 mM HFIP and 16.3 mM TEA concentration (where 16.3 mM TEA was the highest concentration possible to dissolve in 400 mM HFIP at room temperature). Interestingly, they also showed that HFIP effectively disrupts any oligonucleotide secondary structures and this buffer can be an efficient denaturant that allows for more efficient oligonucleotide separations.
As reported by Huber, Oefner, and Bonn (39,51–53), nonporous poly(styrene–divinylbenzene) (PS-DVB) particles with a diameter of 2.1 μm were prepared and successfully used for the ion-pair reversed-phase LC separation of oligonucleotides with methods using 100 mM TEAA as the ion-pairing reagent and a column temperature of 50 °C. Other reversed-phase media such as porous C18 columns can also be used.
The mass transfer of relatively larger molecules such as oligonucleotides is one of the major factors contributing to peak broadening on porous C18 stationary phases. A study using 50 mm × 4.6 mm columns packed with 5-, 3.5-, and 2.5-μm particles showed that the mass transfer in the stationary phase had a major impact on the separation (42). To overcome this problem, columns containing smaller particles (≤2.5 μm in diameter) and using ultrahigh-pressure liquid chromatography (UHPLC) can improve overall separation performance (40). In addition, the use of core–shell C18 particle columns has been shown to significantly improve oligonucleotide separations compared to fully porous particles (54). Systematic evaluation of different core–shell C18 columns showed the best separation with a sub-2-μm core–shell particle column. However, the long-term stability of these silica-based columns when operating at neutral pH and elevated column temperatures of >60 °C can be an issue.
Mixed-Mode Liquid Chromatography: Because of differences in the separation mechanism between the strong-anion-exchange LC and ion-pair reversed-phase LC methods, these approaches are somewhat complementary and often both methods are used for complete analysis and characterization of oligonucleotide mixtures (55,56). As a result, mixed-mode chromatography columns possessing both reversed-phase and ion-exchange properties have been prepared and evaluated for use in oligonucleotide separations (57,58). When using mixed-mode columns, oligonucleotides can experience ionic (such as in strong-anion-exchange LC) and hydrophobic (such as in reversed-phase LC) interactions simultaneously. The dominating mode of interaction can be significantly influenced by the type of mobile phase used.
More recently, an evaluation of different Scherzo C18 mixed-mode (Imtakt USA) columns showed a significant benefit when using mixed-mode columns for separation of oligonucleotides (59). In addition to providing good separation of typical oligonucleotide impurities, such as N–x deletions, which can be well separated by either strong-anion-exchange LC or ion-pair reversed-phase LC alone, these columns also showed an excellent separation of very challenging isomeric oligonucleotides where one single nucleotide base was reversed with its neighbouring base, affording separations that could not be achieved by either strong-anion-exchange LC or ion-pair reversed-phase LC alone. A mixed-mode chromatography separation of oligonucleotides with the Scherzo SM-C18 column and sodium chloride salt gradient is shown in Figure 7.
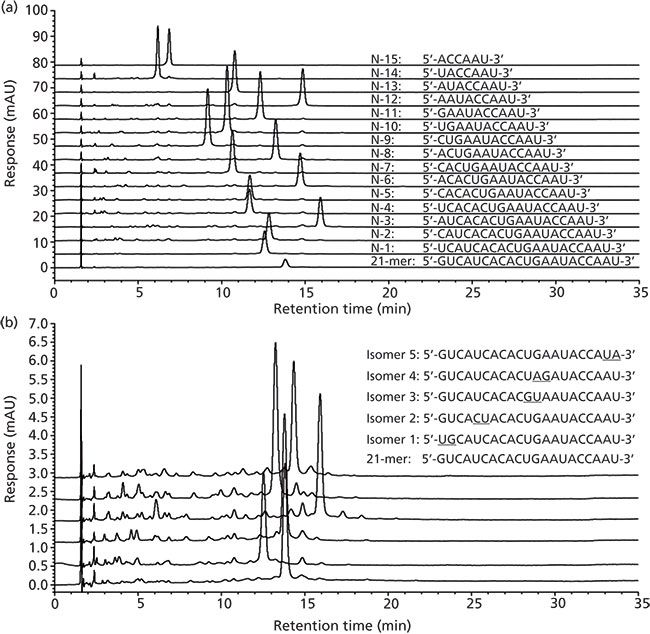
Figure 7: Chromatographic separation of RNA oligonucleotide samples by mixed mode chromatography using a Scherzo SM-C18 column with a sodium chloride gradient: (a) Separation of N–x deletion series, (b) separation of "base-flip" isomer standards. Mobile-phase A: 100 mM Tris (pH 7.4) in water; mobile-phase B: 90:10 (v/v) 2 M sodium chloride in 100 mM Tris (pH 7.4) water–acetonitrile; linear gradient: 50–85% B over 35 min; flow rate: 1 mL/min; column temperature: 50 °C; injection volume: 3 μL; detection: UV absorbance at 260 nm. Adapted with permission from reference 59.
Future Approaches to the Chromatographic Separation of Oligonucleotides
As research on the biomedical uses of short RNA oligonucleotides continues, we can expect ongoing development of improved methods to chromatographically analyze and purify these fascinating compounds. Given the readiness with which these molecules engage in Watson-Crick base pairing with complementary oligonucleotide strands, affinity chromatography-like approaches in which a complementary oligonucleotide stationary phase is used for the targeted retention and chromatographic separation of particular oligonucleotide products may be possible for the analysis, and especially purification, of RNA oligonucleotides. Affinity chromatography purification of oligonucleotide binding proteins has been carried with an oligonucleotide affinity column (oligo (dT)12-18 cellulose) (60) and more recently, with a stationary phase consisting of 2'-fluoro modified RNA covalently linked to agarose beads (61). Oligonucleotide hybridization has long been used in the formation of DNA microarrays and similar technologies (62,63), and can reliably be used for the selective capture of particular oligonucleotide sequences. While these experiments involve only simple binding at lower temperatures with release at elevated temperatures, true chromatographic separation may be possible on complementary oligonucleotide stationary phases operated at elevated temperatures, or in the presence of mobile phase additives that make adsorption or desorption fast on the "HPLC time scale". Such stationary phases could, in principle, be tailor-made for analytical or purification tasks such as the selective binding of desired target oligonucleotides, or of an otherwise difficult-to-remove isomeric or closely related impurity. A preliminary investigation showed that the use of RNA-based stationary phases for selective purification of short RNA sequences suffers from rapid degradation of the RNA stationary phase at elevated column temperatures (64), a problem that could potentially be solved by the use of more thermostable DNA-based stationary phases or even the use of complementary phases based on peptide nucleic acids (PNAs), which are significantly more thermostable than either RNA or DNA, but retain the ability to form complementary duplexes (65).
Conclusions
Developing good chromatographic methods for the accurate and sensitive analysis and separation of oligonucleotides is a critical part in biomedical investigations involving antisense and siRNA oligonucleotides. In this review, we have surveyed different LC approaches including strong-anion-exchange, ion-pair reversed-phase, mixed-mode, and affinity liquid chromatography. Strong-anion-exchange LC is one of the most often used methods in which separation of oligonucleotides is mainly based on the different charges, making this technique especially useful for the separation of the very common N–x deletion impurities. Ion-pair reversed-phase LC is also another commonly used method where, in addition to charge–charge interactions, hydrophobic interactions mainly govern the retention and separation mechanism, making this technique useful for detecting small chemical changes on the full-length sequence. Mixed-mode chromatography, consisting of both reversed-phase and ion-exchange separation modes, provides additional benefits where a single column and method can be used for complete oligonucleotide analysis. Finally, preliminary studies suggest that oligonucleotide-based chromatographic separation may be another useful method for selective separation and purification of these important compounds.
Mirlinda Biba is an Associate Principal Scientist in the Analytical Chemistry Department at Merck in Rahway, New Jersey, USA. She is also a part-time PhD student at Drexel University in Philadelphia, Pennsylvania, USA, studying under the direction of Professor Joe Foley and Dr. Chris Welch. Her research focuses on analysis and separation of short RNA oligonucleotides by different liquid chromatography techniques. Direct correspondence to: mirlinda_biba@merck.com
Joe P. Foley is Professor of Chemistry and Associate Department Head at Drexel University, and a lifetime member of the Chromatography Forum of the Delaware Valley (CFDV). He received his PhD in Chemistry from the University of Florida, Florida, USA, and followed it with a two-year NRC postdoc at NIST. His research interests are in the fundamental and applied aspects of analytical chemistry and separation science, and he has authored or co-authored more than 110 articles, book chapters, reviews, and one patent pertaining to pressure- and voltage-driven liquid-phase chiral and achiral separations (that is, HPLC, UHPLC, SFC, and CE/EKC).
Christopher J. Welch is a science lead for analytical chemistry within the Process and Analytical Chemistry area at Merck Research Laboratories in Rahway. Chris also co-chairs the New Technologies Review and Licensing Committee (NT-RLC), the organization that oversees identification, acquisition, and evaluation of new technologies of potential value to Merck Research Laboratories. Chris also co-chairs the MRL Postdoctoral Research Fellows Programme.
Bing Mao is currently Director, Analytical Chemistry within Process and Analytical Chemistry Department at Merck Research Laboratories in Rahway. He has more than 17 years of experience at Merck with analytical characterization and development to support pharmaceutical small molecule, peptide, and oligonucleotide drug substance manufacturing process development and optimization.
References
(1) L.M. Jarvis, Chem. & Eng. News 87(36), 18–27 (2009).
(2) M.A. Zenkova, D.V. Pyshnyi, W.A. Zalloum, M.B. Gebrezgiabher, and E.V. Bichenkova, Int. Drug Discovery6, 28–32 (2011).
(3) D. Bumcrot, M. Manoharan, V. Koteliansky, and D.W.Y. Sah, Nature Chem. Biol. 2(12), 711–719 (2006).
(4) A. Persidis, Nat. Biotechnol.17, 403–404 (1999).
(5) S.T. Crooke, Antisense Nuc. Acid Drug Dev. 8(2), 115–122 (1998).
(6) T. Tuschl, Chembiochem. 2, 239–245 (2001).
(7) D. Cejka, D. Losert, and V. Wacheck, Clin. Sci. 110, 47–58 (2006).
(8) E. Song, S.K. Lee, J. Wang, N. Ince, N. Ouyang, J. Min, J. Chen, P. Shankar, and J. Lieberman, Nature Med.9(3), 347–351 (2003).
(9) K.V. Morris, S.W. Chan, D.E. Jacobsen, and D.J. Looney, Science305(5688), 1289–1292 (2004).
(10) B.L. Davidson and P.B. McCray, Jr., Nature Rev. Genetics12, 329–340 (2011).
(11) M. Manoharan, Curr. Opin. Chem. Biol.8(6), 570–579 (2004).
(12) J.K. Watts, G.F. Deleavey, and M.J. Damha, Drug Discovery Today13(19/20), 842–855 (2008).
(13) F. Czauderna, M. Fechtner, S. Dames, H. Aygun, A. Klippel, G.J. Pronk, K. Giese, and J. Kaufmann, Nuc. Acids Res.31(11), 2705–2716 (2003).
(14) S.L. Beaucage and M.H. Caruthers, Tetrahedron Lett. 22(20), 1859–1862 (1981).
(15) D. Capaldi, K. Ackley, D. Brooks, J. Carmody, K. Draper, R. Kambhampati, M. Kretschmer, D. Levin, J. McArdle, B. Noll, R. Raghavachari, I. Roymoulik, B.P. Sharma, R. Thurmer, and F. Wincott, Drug Inf. J.46, 611–626 (2012).
(16) S.P. Waghmare, P. Pousinis, D.P. Hornby, and M.J. Dickman, J. Chromatogr. A1216, 1377–1382 (2009).
(17) M.B. Beverly, Mass Spectrom. Rev. 30, 979–998 (2011).
(18) A. Andrus, Methods: A companion to Methods in Enzymol.4, 213–226 (1992).
(19) F.H.M. van Dinther, R.W. Bally, and T.P.G. van Sommeren, J. Chromatogr. A817, 273–279 (1998).
(20) K. Cook and J. Thayer, Bioanalysis3(10), 1109–1120 (2011).
(21) M. Dizdaroglu and W. Hermes, J. Chromatogr. 171, 321–330 (1979).
(22) J.H. van Boom and J.F.M. de Rooy, J. Chromatogr. 131, 169-177 (1977).
(23) W. Haupt and A. Pingoud, J. Chromatogr. 260, 419–427 (1983).
(24) W. Jost and K.K. Unger, J. Chromatogr.185, 403–412 (1979).
(25) J.D. Pearson and F.E. Regnier, J. Chromatogr.255, 137–149 (1983).
(26) T.G. Lawson, F.E. Regnier, and H.L. Weith, Anal. Biochem. 133, 85–93 (1983).
(27) R.R. Drager and F.E. Regnier, Anal. Biochem.145, 47–56 (1985).
(28) H.A. Claessens, M.A. van Straten, and J.J. Kirkland, J. Chromatogr. A 728, 259–270 (1996).
(29) C. McNeff and P.W. Carr, Anal. Chem.67, 2350–2353 (1995).
(30) Y. Kato, T. Kitamura, A. Mitsui, and T. Hashimoto, J. Chromatogr.398, 327–334 (1987).
(31) Y. Kato, T. Kitamura, A. Mitsui, Y. Yamasaki, and T. Hashimoto, J. Chromatogr. 447, 212–220 (1988).
(32) J.R. Thayer, V. Barreto, S. Rao, and C. Pohl, Anal. Biochem. 338, 39–47 (2005).
(33) A. Sabarudin, J. Huang, S. Shu, S. Sakagawa, and T. Umemura, Anal. Chim. Acta 736, 108–114 (2012).
(34) M. Buncek, V. Backovska, S. Holasova, H. Radilova, M. Safarova, F. Kunc, and R. Haluza, Chromatographia62, 263–269 (2005).
(35) D.G. Volkmann, Anal. Chem. Progress126, 51–69 (1984).
(36) C. Horvath, W. Melander, I. Molnar, and P. Molnar, Anal. Chem.49, 2295–2305 (1977).
(37) R.P.W. Scott and P. Kucera, J. Chromatogr. 175, 51–63 (1979).
(38) P.J. Oefner, J. Chromatogr. B739, 345–355 (2000).
(39) C.G. Huber, P.J. Oefner, and G.K. Bonn, Anal. Biochem. 212, 351–358 (1993).
(40) S.M. McCarthy, M. Gilar, and J. Gebler, Anal. Biochem.390, 181–188 (2009).
(41) H. Oberacher, A. Krajete, W. Parson, and C.G. Huber, J. Chromatogr. A 893, 23–35 (2000).
(42) M. Gilar, K.J. Fountain, Y. Budman, U.D. Neue, K.R. Yardley, P.D. Rainville, R.J. Russell II, and J.C. Gebler, J. Chromatogr. A 958, 167–182 (2002).
(43) A. Apffel, J.A. Chakel, S. Fischer, K. Lichtenwalter, and W.S. Hancock, Anal. Chem.69, 1320–1325 (1997).
(44) M. Gilar, Anal. Biochem.298, 196–206 (2001).
(45) S. Li, D.D. Lu, Y.L. Zhang, and S.Q. Wang, Chromatographia 72, 215–223 (2010).
(46) B. Noll, S. Seiffert, H.P. Vornlocher, and I. Roehl, J. Chromatogr. A1218, 5609–5617 (2011).
(47) K.J. Fountain, M. Gilar, and J.C. Gebler, Rapid Commun. Mass Spectrom.17, 646–653 (2003).
(48) V.B. Ivleva, Y.Q. Yu, and M. Gilar, Rapid Commun. Mass Spectrom.24, 2631–2640 (2010).
(49) M. Beverly, K. Hartsough, L. Machemer, P. Pavco, and J. Lockridge, J. Chromatogr. B 835, 62–70 (2006).
(50) P. Deng, X. Chen, G. Zhang, and D. Zhong, J. Pharm. Biomed. Anal.52, 571–579 (2010).
(51) C.G. Huber, P. J. Oefner, E. Preuss, and G. K. Bonn, Nuc. Acids Res.21(5), 1061–1066 (1993).
(52) C.G. Huber, P.J. Oefner, and G.K. Bonn, Anal. Chem.67, 578–585 (1995).
(53) C.G. Huber, E. Stimpf, P.J. Oefner, and G.K. Bonn, LCGC North Am. 14(2), 114–127 (1996).
(54) M. Biba, C.J. Welch, J.P. Foley, B. Mao, E. Vazquez, and R.A. Arvary, J. Pharm. Biomed. Anal.72, 25–32 (2013).
(55) T. Kremmer, M. Boldizsar, and L. Holczinger, J. Chromatogr. 415, 53–63 (1987).
(56) L.W. McLaughlin, J. Chromatogr.418, 51–72 (1987).
(57) R. Bischoff and L.W. McLaughlin, Anal. Biochem. 151, 526–533 (1985).
(58) L.W. McLaughlin, Chem. Rev.89, 309–319 (1989).
(59) M. Biba, E. Jiang, B. Mao, D. Zewge, J.P. Foley, and C.J. Welch, J. Chromatogr. A 1304, 69–77 (2013).
(60) S. Okamura, F. Crane, H.A. Messner, and T.W. Mak, J. Biol. Chem.253(11), 3765–3767 (1978).
(61) R.H. Hovhannisyan and R.P. Carstens, BioTechniques 46, 95–98 (2009).
(62) E.A. Smith, M. Kyo, H. Kumasawa, K. Nakatani, I. Saito, and R.M. Corn, J. Am. Chem. Soc.124, 6810–6811 (2002).
(63) F. Seela and S. Budow, Mol. BioSyst.4, 232–245 (2008).
(64) M. Biba, C.J. Welch, and J.P. Foley, unpublished data (2013).
(65) Y. Aiba, Y. Hamano, W. Kameshima, Y. Araki, T. Wada, A. Accetta, S. Sforza, R. Corradini, R. Marchelli, and M. Komiyama, Org. & Biomol. Chem.11(32), 5233–5238 (2013).


.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)













