Analysis of D-Amino Acids: Relevance in Human Disease
Small metabolic molecules often are chiral and can play important roles in regulating a variety of biological functions, occasionally providing information about the presence and progression of disease. The most ubiquitous class of such small molecules are amino acids. Sensitive and accurate analysis of the less prevalent D-amino acids as free entities or as constituents of peptides can be challenging, particularly when complex physiological matrices are involved. The number of studies involving low-abundance D-amino acids in biological systems has increased significantly over the last decade. Studies involving their presence and importance have become increasingly difficult to ignore. Their relevance in neurological pathologies, cancer, kidney disorders, and more, has advanced. Chiral separations have played and continue to play a central role in these studies. Because enantiomers and epimers have the same exact mass, stereoselective separations are essential. However, sensitive detection is also necessary because trace levels of these analytes are involved. Multidimensional separations often provide the best avenue for accurate qualitative and quantitative results. Future developments will involve faster and highly specific routine testing, particularly if these analyses are to enter the clinical realm.
Specific small-molecule metabolic biomarkers can be essential tools for detecting and diagnosing diseases and monitoring their progress. Furthermore, they can serve as signaling and activation molecules and can even play a role in developing new therapeutics. Many small-molecule biomarkers are chiral and the dominant class of these are amino acids. Most proteinic and non-proteinic amino acids exist in the dominate L-enantiomeric form; the “D-form” is less common. Studies on both types of D-amino acids (proteinic and nonproteinic) in human disease have increased significantly over the last decade. Samples of various human fluids and tissues can be limited in amounts, involve complex matrices, and often have only trace levels of D-amino acids or epimeric peptides. Because enantiomers and epimeric peptides have the same exact mass, separations play the dominant role in their analysis. However, mass spectrometry (MS) can be a highly useful detector, given its sensitivity. Fluorescence detection is equally or more sensitive but does not provide m/z analyte corroboration, which can be useful in complex samples with overlapping peaks (1). In both cases, detection can benefit from analyte derivatization, which also aids with sample preparation and preconcentration (1,2).
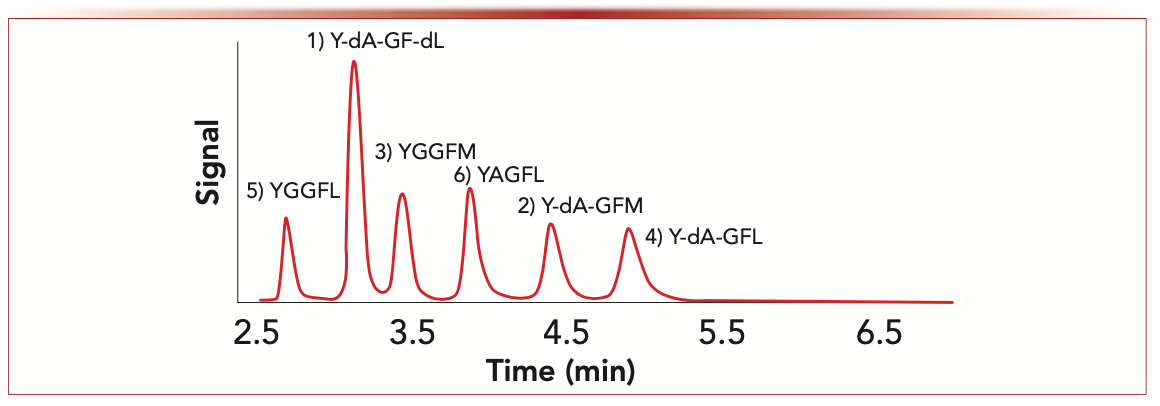
Multidimensional separations can be highly useful to minimize matrix effects for complex biological samples where multiple amino acids or peptides are to be determined in a single analysis. Given that D-amino acids in physiological samples nearly always exist in the presence of much higher levels of the L-amino acid enantiomers, good chiral separations are necessary (3,4). High performance liquid chromatography (HPLC) is the dominant approach for such analyses. Gas chromatography (GC) and capillary electrophoresis (CE) have also been used for this type of analysis, albeit to a much lesser extent. Even when analyzing peptide epimers, using chiral stationary phases often provides superior results, as shown in Figure 1 (5,6).
FIGURE 1: Enkephalin peptide stereoisomers: separation of enkephalin peptide epimers on a 100 mm x 4.6 mm, i.d. TeicoShell superficially porous particle (SPP) chiral column (Azyp, LLC, Arlington, Texas) with UV detection and a flow rate of 1.0 mL/min. Mobile phase = 65:35 (v:v) acetonitrile:aqueous 2.5 mM ammonium formate (NH4HCO2).
The acceptance of the physiological role and the importance of the diagnostic value of D-amino acids in human disease (and mammals, in general) was slow to materialize even though their importance as essential elements in the cell wall peptidoglycan of bacteria was well known (7). The mainstay scientific and biomedical communities were heavily invested in more routine areas of study and were not anxious to have investigations in areas they did not understand or for which they did not have facile, readily available methodologies at their disposal. This attitude began to change at the turn of the century. An important ion channel in neurological tissue, referred to as N-methyl- D-aspartate (NMDA) receptors, began to shift the attitudes of some scientists. In 2000, it was reported that D-serine was an important agonist for NMDA receptors (8). Subsequent to the discovery of D-serine as an NMDA agonist, D-aspartic acid and D-alanine were found to function similarly. NMDA receptors also were found to be upregulated in cancer cells (9–11). Over the last decade, studies on D-amino acids in biological systems have increased exponentially.
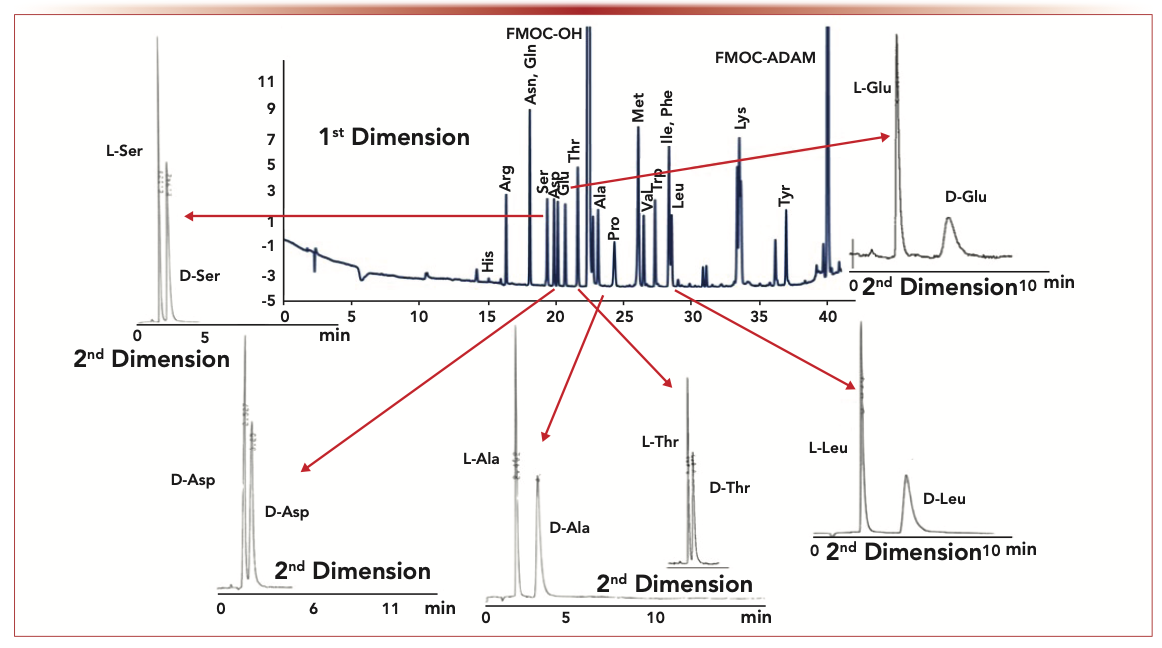
It is interesting to note that Nobel Laureate H.A. Krebs discovered the existence of D-amino acid oxidase in mammals in 1935 (12). Two things are notable about this relatively ignored discovery. First, this report predated any publications on the existence of naturally occurring D-amino acids in mammals or even in bacteria. Second, few, if any, biographies, reviews, or synopses of Krebs’s substantial and important body of work ever mention this seminal discovery. Our early work involving D-amino acids involved the HPLC enantiomeric chromatographic separation of dansyl-derivatized amino acids on cyclodextrin bonded chiral stationary phases (CSPs) that we first developed (13). Interestingly, the columns were commercialized in 1983, but the first paper didn’t appear until 1984 given patent and review matters. A 1991 study showed that D-amino acids were prevalent in human urine (14). There were a number of other important ramifications of this work. It demonstrated the importance of multidimensional LC (MDLC) for analyzing enantiomers in biological samples, which in this case was achiral-chiral, heart-cutting two-dimensional (2D)-LC. Furthermore, this study supported the contention that there were no enantiomerically pure proteinic amino acids—either biological or commercial. Finally, the superiority of chromatography for determining enantiomeric excess was (and is) unsurpassed. For example, the detection of less than one part per 100,000 of an enantiomer was clearly demonstrated. By 1993, the ubiquitous presence of D-amino acids in human plasma, cerebrospinal fluid, and amniotic fluids was known (15). It was shown that some biological fluids were best analyzed by three-dimensional (3D)-LC (achiral-achiral-chiral). Effective analytical methodologies were critical for good qualitative and quantitative determination of typically trace levels and often variable D-amino acid amounts in biological samples. Questions regarding the origins and relevance of these antipodes have been asked continually, and the search for answers is escalating today. In mammals, there are at least three important sources of D-amino acids, which are food intake, gut bacteria, and intrinsic biosynthesis (16). The latter of these may be the most intriguing in terms of signaling molecules, biomarkers for disease, and so on. Early on, we developed a 2D-LC method with tandem UV and fluorescence detection of N-(fluorenyl-9-methoxycarbonyl) amino acids (FMOC-AAs) to evaluate these factors in the plasma and urine of laboratory rodents (see Figure 2). Among the interesting results was that pregnancy had a profound effect on the excretion of D-amino acids (16). This same analytical approach was used to show that excretion levels of a nonproteinic D-amino acid (pipecolic acid) could be directly correlated with the severity of a family of genetic metabolic diseases, referred to as peroxisomal deficiencies (17). The more severe disorders of this type are invariably fatal and are preceded by severe retardation. Subsequently, it was reported that high concentrations of certain D- and L-amino acids could inhibit the growth of Chinese hamster ovary (CHO) cells and HeLa cells (18). This effect was thought to be because of the production of hydrogen peroxide (H2O2) from D-amino acid oxidase and L-amino acid oxidase. The essential determination of lipid peroxidation was done by HPLC with fluorescence detection.
FIGURE 2: Achiral-chiral heart-cutting using 2D-LC: In this separation sequence of analytes, the first dimension utilized a gradient reversed-phase separation C18 SPP (Poroshell 120 EC-C18, 4.6 × 150 mm, and the second dimension utilized a TeicoShell SPP CSP (4.6 x 100 mm). Flow rates were both 1.0 mL/min. UV detection was used in the first dimension and fluorescence detection was used in the second dimension. Axes labels are retention time (min) for the x-axis and detector signal intensity for the y-axis.
It was found that high levels of D-serine can overstimulate NMDA receptors and may be implicated in a number of neurodegenerative diseases. Additionally, D-amino acid–containing peptides (epimers) were found in the central nervous system and may also be implicated in neurological disorders, including Alzheimer’s disease (19–21). In a groundbreaking study, D-leucine was found to have potent antiseizure effects in humans (22). Given the prevalence and importance of NMDA receptors in the central nervous system, it was not surprising to find that specific D-amino acids were prevalent in various brain tissues (3). In many of these studies, 2D-LC played a fundamental role. Among the more interesting results were that D-glutamic acid was the least prevalent D-amino acid in brain tissue. There appears to be an active removal mechanism for D-glutamic acid. Could this be connected to the fact that L-glutamic acid is a potent and prevalent neurotransmitter?
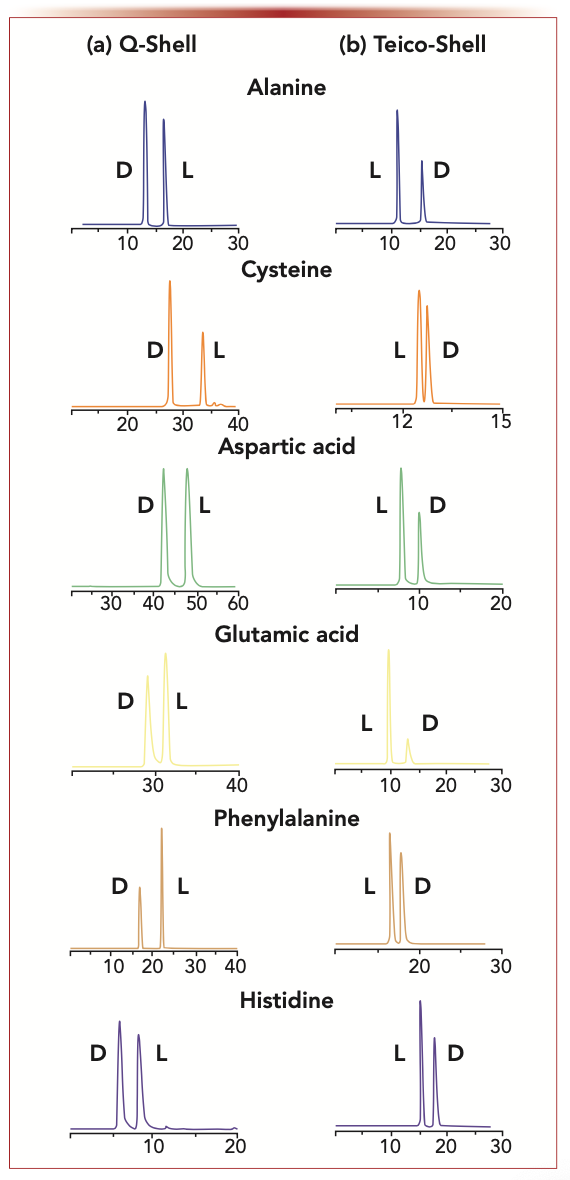
Given that NMDA receptors are up-regulated in many types of cancer cells (9–11), it is not surprising that D-amino acids are of growing importance as biomarkers and possibly in therapeutic strategies. Both 2D-LC (achiral-chiral) and chiral LC with tandem mass spectrometry (LC–MS/MS) have been indispensable tools in such studies. Indeed, CSPs that can reverse the elution order of amino acid enantiomers are often essential tools for obtaining accurate qualitative and quantitative results, as shown in Figure 3. Using such tools, it was found that cancer cells quickly released D-alanine and D-aspartic acid when their viability was diminished by the presence of NMDA blocking agents (11). The released D-amino acids appeared to counteract the effects of the blocking agents, thereby “revitalizing” the cancer cells.
FIGURE 3: Reversal of the enantiomeric elution order of amino acids: complete reversal of enantiomeric elution order of representative 6-aminoquinolyl-carbamyl (AQC)-amino acids on a 150 x 3 mm i.d. TeicoShell and Q-Shell chiral stationary phases (Azyp, LLC). This reversal occurs for all tested chiral native or N-blocked amino acids. (a) Q-Shell conditions: flow rate= 0.4 mL/min, gradient; mobile phase A; 90:10 methanol: 50 mM ammonium formate pH 6, mobile phase B; 90:10 methanol:50 mM ammonium formate pH 4.0, 0–12 min 0% B, 12–17 min 0–100% B, 17–55 min 100% B, 55–60 min 100–0% B. (b) TeicoShell conditions: flow rate = 0.4 mL/min, gradient; mobile phase A; 5 mM ammonium formate pH 4, mobile phase B; acetonitrile with 0.1% formic acid, 0–3 mins 5% B, 3–18 mins 5–35% B, 18–20 mins 35–50% B, 20–23 min 50–80% B, 23–39 min 80% B, and 39–40 min 5% B. Axes labels are retention time (min) for x-axis and detector signal intensity for y-axis.
It has been shown that high D-serine levels can have detrimental effects on the kidney. Conversely, low D-serine levels can exacerbate ischemia and reperfusion injuries. A 2D-LC–MS/MS system was used for chiral amino acid metabolomics for screening and prognosis of chronic kidney disease (CKD) (23,24). D-aspartic acid and D-proline were associated with diabetes mellitus, whereas D-serine and D-aspartate were strongly associated with the progression of CKD.
The prevalence and role of D-amino acids in human diseases continues to expand into unexpected areas. For example, nonproteinic aminobutyric acids have been shown to have clinical applications as biomarkers for osteoporosis (25). Again, chiral LC–MS/MS with a TeicoShell column was the fundamental technique in analyzing the biological tissues. It was shown that (R)–aminoisobutyric acid in serum was positively associated with hip bone mineral density in older individuals. Recently, D-cysteine was shown to be an endogenous regulator of neural progenitor cell dynamics in the mammalian brain (26). Chiral LC–MS and a stereospecific luciferase assay were used to identify endogenous D-cysteine in the brain. D-cysteine was enriched more than 20-fold in the embryonic brain.
Clearly, the importance of analyzing D-amino acids as single entities and as constituents of peptides and proteins is expanding enormously. The essential analytical methodologies that have been developed will continue to improve and be a central part of the discovery process. They will be essential tools that allow a fundamental understanding of the mechanism of action and biological role of the often “misunderstood” D-amino acids. High-throughput screening will require fast separations if such analyses are to move to the clinical setting (27,28).
References
(1) Y.S. Sung, A. Berthod, D. Roy, and D.W. Armstrong, Chromatographia 84, 719–727 (2021).
(2) Y. Sung, J. Putman, S. Du, and D.W. Armstrong, Anal. Biochem. 642, 114451 (2022)
(3) C.A. Weatherly, S. Du, C. Parpia, P.T. Santos, A.L Hartman, and D.W. Armstrong, ACS Chem. Neuro. 8, 1251–1261 (2017).
(4) S. Du, Y. Wang, C.A. Weatherly, K. Holden, and D.W. Armstrong, Anal. Bioanal. Chem. 410, 2971–2979 (2018).
(5) S. Du, E.R. Readel, M. Wey, and D.W. Armstrong, Chem. Commun. 56, 1537–1540 (2020).
(6) R.M. Wimalasinghe, Z.S. Breitbach, J.T. Lee, and D.W. Armstrong, Anal. Bioanal. Chem. 409, 2437–2447 (2017).
(7) J.T. Park, J. Biol. Chem. 194, 897–904 (1952).
(8) J.D. Mothet, A.T. Parent, H. Wolosker, R.O. Brady, Jr., C.D. Ferris, M.A. Rogaski, and S.H. Snyder, Proc. Nat. Acad. Sci. 97, 4926–4931 (2000).
(9) A. Stepulak, H. Luksch, C. Gebhardt, O. Uckermann, J. Marzahn, M. Sifringer, et al, Histochem. Cell Biol. 132, 435–445 (2009).
(10) S.I. Deutsch, A.H. Tang, J.A. Burket, and A.D. Benson, Biomed. Pharmacother. 68, 493–496 (2014).
(11) S. Du, Y.S. Sung, M. Wey, Y. Wang, N. Alatrash, A. Berthod, et al, Mol. Biol. Rep. 47(9), 6749–6758 (2020).
(12) H.A. Krebs, Biochem. J. 29, 1620–1644 (1935).
(13) D.W. Armstrong and W. DeMond, J. Chromatogr. Sci. 22, 411–415 (1984).
(14) D.W. Armstrong, J.D. Duncan, and S.H. Lee, Amino Acids 1, 97–106 (1991).
(15) D.W. Armstrong, M.P. Gapser, S.H. Lee, J. Zukowski, and N. Ercal, Chirality 5, 375–378 (1993).
(16) D.W. Armstrong, M.P. Gasper, S.H. Lee, N. Ercal, and J. Zukowski, Amino Acids 5, 299–315 (1993).
(17) D.W. Armstrong, J. Zukowski, N. Ercal, and M.P. Gasper, J. Pharm. Biomed. Anal. 11, 881–886 (1993).
(18) N. Ercal, X. Luo, R.H. Matthews, and D.W. Armstrong, Chirality 8, 24–29 (1996).
(19) S.D. Heck, C.J. Siok, K.J. Krapcho, P.R. Kelbaugh, P.F. Thadeio, M.J. Welch, et al, Science 266, 1065–1068 (1994).
(20) T. Kubo, Y. Kumagae, C.A. Miller, and I. Kaneko, J. Neuropathol. Exp. Neurol. 62, 248–259 (2003).
(21) S. Du, E.R. Readel, M. Wey, and D.W. Armstrong, Chem. Comm. 56, 1537–1540 (2020).
(22) A.L. Hartman, P. Santos, K.J. O’Riordan, C.E. Statfstrom, and M.J. Hartwick, Neurobiol. Dis. 82, 46–53 (2015).
(23) G. Young, S. Kendall, and A. Brownjohn, Amino Acids 6, 283–293 (1994).
(24) T. Kimura, et al, Sci. Rep. 6, 1–7 (2016).
(25) Z. Wang, L. Brian, C. Mo, H. Shen, L.J. Zhao, K.J. Su, et al, Commun. Biol. 3(39), s42003-020-0766-y (2020).
(26) E.R. Semenzaa, M.M. Harraza, E. Abramson, A.P. Mallaa, C. Vasavdaa, M.M. Gadalla, et al, Proc. Nat. Acad. Sci. 118, e2110610118 (2021).
(27) D.C. Patel, Z.S. Breitbach, M.F. Wahab, C.L Barhate, and D.W. Armstrong, Anal. Chem. 87, 9137–9148 (2015).
(28) C.L. Barhate, E.L. Regalado, N.D. Contrella, J. Lee, J. Jo, A.A. Makarov, et al, Anal. Chem. 89, 3545–3553 (2017).
Daniel W. Armstrong is the Welch Distinguished Professor with the Department of Chemistry & Biochemistry at the University of Texas at Arlington, Texas. Direct correspondence to: sec4dwa@uta.edu.


Regulatory Deadlines and Supply Chain Challenges Take Center Stage in Nitrosamine Discussion
April 10th 2025During an LCGC International peer exchange, Aloka Srinivasan, Mayank Bhanti, and Amber Burch discussed the regulatory deadlines and supply chain challenges that come with nitrosamine analysis.