Simplifying Peptide Bioanalysis
Biomolecules, such as proteins and peptides, represent a growing number of pharmaceutical drug entities.
Erin E. Chambers, Jessalynn Wheaton, and Diane M. Diehl, Waters Corporation
Biomolecules, such as proteins and peptides, represent a growing number of pharmaceutical drug entities. These biopharmaceuticals are often more specific than small molecules, since they typically replace or augment an endogenous molecule known to change a disease or physiological state (1). Since they are analogs to or synthetic versions of endogenous compounds, they are usually well tolerated by the body. There are currently over 40 peptide-based drugs on the market and over 400 in late stage clinical trials (2).
Analytical Challenges
Reliable, robust, sensitive, and accurate bioanalytical methods need to be developed for biopharmaceuticals, as well as for small molecules. Working with peptides presents many additional challenges over those typically faced with small molecules. Solubility and adsorption pose serious complications during method development. One must pay particular attention to the organic concentration and modifier content of the injection solvent, sample preparation elution solvent, and any LC wash solvents. Problems with adsorption make sample concentration (which is often needed in order to meet detection limits) without evaporation highly desirable. Obtaining selectivity for peptide therapeutics in complex biological matrices is also crucial. Selective sample prep continues to play a critical role in minimizing matrix effects and reducing variability associated with incurred samples. Because of their size and charge state distribution, sensitivity by mass spectrometry may be lower for biomolecules than typical small molecules.
Solution
The Peptide Separation Technology (PST) Method Development Kit was designed to facilitate the development of selective extraction and liquid chromatographic methods for peptides in biological matrices. The PST Method Development Kit is available for ACQUITY UPLC® (part#176001835) or HPLC (part#176001836). The kit contains a chromatographic column, the Oasis® μElution PST Method Development extraction plate and simple, straightforward SPE and LC screening protocols. The kit was used to develop extraction and chromatographic methods for the 12 peptides described in Table I.
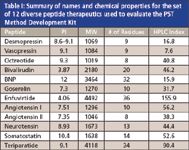
Table I: Summary of names and chemical properties for the set of 12 diverse peptide therapeutics used to evaluate the PST Method Development Kit
Experimental
Spiked human plasma samples were pre-treated by diluting 1:1 (v:v) with 4% H3PO4 in H2O. Samples were then extracted using the Oasis® μElution PST Method Development extraction plate according to the generic screening protocol included in the kit. An ACQUITY UPLC® system was used to increase sensitivity, resolution, and separation speed. Chromatographic separation of peptides was performed with the UPLC PST column and generic screening protocol included in the kit. A Xevo™ TQ MS was used to detect the peptides in positive ion electrospray. The mass range on both quadrupoles of the Xevo™ TQ MS is m/z 2048, a key MS characteristic when one considers that the most intense precursor masses for some of the larger peptides were present at m/z values between 1200 and 1500. As UPLC peak widths are on the order of 2–3 s wide at base for all peptides, it is also important to have a mass spectrometer capable of providing enough points across the peaks for accurate and reproducible quantitation. LC–MS performance, using the generic LC screening protocol from the kit is shown in Figure 1. It should be noted that a critical pair, vasopressin and desmopressin (differing only by an amino group), are well separated using the generic LC method.

Figure 1
Results and Discussion
In the extraction solution provided in the PST kit, the orthogonal nature of mixed-mode solid phase extraction (SPE) is used to increase selectivity over conventional methods, since it relies on both reversed-phase and ion exchange mechanisms. Mixed-mode SPE selectively separates the desired analytes from matrix components and has been shown to reduce matrix effects (3). Matrix effects, where measured, were <11% for all peptides, indicating specificity of the methodologies. The screening protocol yielded >82% recovery for 9 out of 12 peptides extracted from human plasma. Minor modifications to the generic screening protocol improved recovery for the other 3 peptides to >83%. Figure 2 summarizes SPE recovery for the test set of 12 peptide therapeutics extracted using the SPE screening protocol and plate from the kit. The extraction was performed in 96-well Oasis® μElution plate format to obtain the analyte concentration necessary to meet required LOD/LLOQs without evaporation and reconstitution, steps that often lead to peptide losses. A concentration factor of up to 15× can be achieved without evaporation using the μElution format. Figures 3 and 4 demonstrate representative LLOQs for peptides extracted from human plasma using the generic LC and SPE screening protocols without optimization.
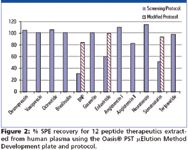
Figure 2
Conclusions
The Oasis® PST μElution Method Development plate and protocol can greatly simplify extraction method development and rapidly identifies an appropriate SPE sorbent and selective starting protocol for peptide bioanalysis. Peaks widths on the order of 2–3 s wide at base, and >15 points across the peak, were obtained for all peptides tested using the ACQUITY UPLC® PST column provided in the kit. The Xevo™ TQ MS system provided the mass range, scan speed, and sensitivity required for peptide bioanalysis.
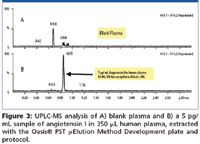
Figure 3
Overall, we have shown that bioanalytical studies for peptide therapeutics are amenable to a platform-based approach to methods development. Such standardized approaches for determining optimal SPE recovery and concentration, and MRM-based LC–MS analysis should permit companies to reduce development timelines and shorten time to market for their peptide drugs.
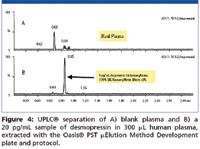
Figure 4
For the complete UPLC application note, visit www.waters.com/3253
References
(1) Bladon, C., Pharmaceutical Chemistry: Therapeutic Aspects of Biomolecules, Wiley and Sons, West Sussex, England, 2002.
(2) Bioscience Technology, January 2009.
(3) Chambers, E., Wagrowski-Diehl, D. M., Lu, Z., Mazzeo, J. R. J. Chromatogr. B 2007, 852 (1–2), 22–34.
© 2010 Waters, ACQUITY UPLC, UPLC are registered trademarks of Waters Corporation. Xevo and The Science of What's Possible are trademarks of Waters Corporation. All other trademarks are the property of their respective owners.

Waters Corporation
34 Maple Street, Milford, MA 01757
tel. (508)478-2000; fax (508)482-3605
Website: www.waters.com

SEC-MALS of Antibody Therapeutics—A Robust Method for In-Depth Sample Characterization
June 1st 2022Monoclonal antibodies (mAbs) are effective therapeutics for cancers, auto-immune diseases, viral infections, and other diseases. Recent developments in antibody therapeutics aim to add more specific binding regions (bi- and multi-specificity) to increase their effectiveness and/or to downsize the molecule to the specific binding regions (for example, scFv or Fab fragment) to achieve better penetration of the tissue. As the molecule gets more complex, the possible high and low molecular weight (H/LMW) impurities become more complex, too. In order to accurately analyze the various species, more advanced detection than ultraviolet (UV) is required to characterize a mAb sample.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)














