Biotechnology Highlights from Isranalytica 2011
LCGC North America
Isranalytica offered great talks on biotechnology, on topics from comprehensive protein characterization to middle-out proteomics to capillary electrophoresis for glycoprofiling.
Isranalytica, held this year in Tel Aviv, Israel, on February 7 and 8, is one of the largest analytical chemistry conferences in the world. Here, we bring you highlights of the excellent biotechnology talks we heard there.
Isranalytica 2011, the 14th annual conference organized and administered by the Israel Analytical Chemistry Society, was one of the largest analytical chemistry conferences in the world. Approximately 3000 participants attended, including scientists from academia, government organizations, and the pharmaceutical, chemical, petrochemistry, food, biochemistry, and biological industries, as well as water experts, engineers, quality managers, metrologists, and private contractors. Isranalytica has been recognized by the Analytical Chemistry Division of the European Association for Chemical and Molecular Sciences, the International Society of Electrochemistry, and the Cooperation for International Traceability in Analytical Chemistry.
The program and abstracts for the 2011 meeting can be found at http://Isranalytica.org.il/index.htm (double click on the titles and abstracts will appear). Isranalytica brings together leading companies involved in analytical instrumentation and equipment, bridging technology developments with end users (the conference attendees). The programming for Day 1 consisted of a combination of oral and poster presentations, as well as a special plenary workshop with K. Russo of the United States Pharmacopeia (USP), which was held in the afternoon. A related special session was held on Day 2, entitled, "Meet the Regulator and the Pharmacopeia." Answering questions and hosting the dialog with K. Russo was S. Kozlowski of the U.S. Food and Drug Administration.
This column is devoted to the coverage of all papers, oral or poster, given at this meeting that would be of interest to readers of "Biotechnology Today." Such sessions included Peptides and Protein Analysis, Pharmaceutical Analysis, Mass Spectrometry, Separations, Spectroscopy, Validation, Data Processing, and Patents.
Comprehensive Characterization of Proteins
B.L. Karger opened the conference with a plenary lecture entitled "Comprehensive Characterization of Protein Targets and Biopharmaceuticals," which summarized, in part, his background in collaborating with various Israeli academic centers, the history of the Barnett Institute (BI) at Northeastern University (Boston, Massachusetts), and his ongoing research in biotechnology and protein characterizations. A major thrust at BI currently involves a new emphasis in its new Center for Advances in Regulatory Analysis (CARA). This venture is designed to assist the biotechnology industry in meeting biopharmaceutical regulatory science requirements by means of service and research, all involving good laboratory practices (GLP) and facilities. One of the main goals of CARA is to develop new and improved analytical methods to serve the biotechnology industry, especially through collaborations with firms in the greater Boston area.
High Resolution Mass Spectrometry with CID, ETD, MS2, and MS3
Karger also described the most recent research efforts at BI involving improved characterizations of biotechnology products, especially antibodies, fusion antibodies, and other glycoproteins. Both liquid chromatography–mass spectrometry (LC–MS) and capillary electrophoresis (CE)–MS continue to play critical roles in protein characterization. In recent studies, Karger, with research faculty member Billy Wu, has looked at newer methods to determine higher order structures, such as specific disulfide linkage analyses. This work has involved high-resolution mass spectrometry (HRMS), using a combination of collisionally induced dissociation (CID) and electron transfer dissociation (ETD) together with MS2 and MS3 analyses of proteins as complex as tissue plasminogen activator (tPA) (17 disulfides) (Figure 1; please see the print version of this issue for this figure) (1,2). These techniques have, for the first time, allowed for the determination of the precise location of all disulfides within the tPA structure, and correctly identified the unbound cysteine residue, confirming the original assignments made in earlier work at Genentech. Figure 2 (please see the print version of this issue for this figure) illustrates one of many LC–MS-MS assays of disulfide-linked peptides obtained by means of peptide mapping of intact tPA, showing how such bonds can be located and specified (1,2). In this work, both ETD and CID-MS3 techniques were applied to the assignments of each disulfide bond location and linkages within such peptides. Such approaches are the very latest MS techniques to make possible unambiguous determination of total structures and post-translational modifications (PTMs) present for complex proteins, such as tPA, antibodies, and other glycoproteins.
Capillary Electrophoresis to Analyze for Deamidation and Aspartic Acid Isomerization and Glycoprofiling
Further studies, now in collaboration with Sunny Zhou, a faculty fellow at BI, have involved the development of newer ways to analyze for deamidation and aspartic acid isomerization by means of the analysis of isoaspartic acid formed in the deamidation process using LC–MS with ETD. Intact protein analyses for dozens of isoforms in glycoproteins have been quantitated and studied by CE–MS methods.
Finally, Karger described work pursued together with Andras Guttman, a research faculty member, involving the use of CE to determine glycoprotein variants, glycan analyses, and glycoprofiling, all done using commercially available tagging reagents and methods (Beckman Coulter, Brea, California). This talk was perhaps the most comprehensive overview of what newer analytical techniques are now available, often commercially, to meet biopharmaceutical regulatory science requirements for filing biotechnology applications with the regulatory agencies in the United States and elsewhere.
Proteomics and Protein Sequencing
Krull and Kreimer, from Northeastern University's Chemistry and Chemical Biology Department, presented a keynote lecture on Monday morning entitled "Top-Down vs. Bottom-Up vs. Middle-Out, Shotgun Proteomics Problems and Top-Down Protein Sequencing (TDS)." In this talk, which dealt with modern characterization methods by MS, an overview of the most important protein identification MS methods available today in all of proteomics or peptidomics was presented (3). Krull provided an overview of the latest and past MS-based analytical methods used in proteomics, and their advantages and limitations. Also explained were some of the latest methods, such as TDS, or middle-down, which is really a variant of bottom-up methods, forming larger peptides than with trypsin. It was made clear that bottom-up (or middle-down for that matter), depends entirely on determining peptides found in given proteins, and then making use of online databases to identify the most likely structures for the possible proteins in the proteome. Such approaches then use those peptides formed by peptide mass fingerprinting (PMF) and peptide mapping routines. Whereas bottom-up has used trypsin almost entirely as the enzyme of choice, middle-down uses other enzymes, such as endo-lys-C, that form larger peptides, often more easily located and then sequenced online in LC–electrospray ionization (ESI)–MS-MS routines. The significant disadvantage of both of these techniques, in the absence of top-down MS to determine the intact molecular weight of each and every protein being identified by PMF and peptide mapping, lies in the inability of the database to know whether all relevant peptides were located and identified. And the absence of evidence is not evidence of absence.
Thus, results are almost always based on partial, often incomplete, peptide analyses, and extrapolation to the best possible known, and possibly incorrect, proteins in that specific proteome. Hence, some of the current and future proteomics literature may be questionable, in one form or another. Even middle-down proteomics, in the absence of top-down molecular weight determinations (termed top-down proteomics, which is not TDS), are really variations on bottom-up or shotgun methods, which can and do lead to incorrect protein identifications of unknown extents in literature.
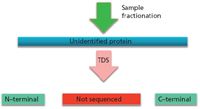
Figure 3: Middle-down protein sequencing (9).
Perhaps the solution to the above problems, which are also related to determining recombinant protein structures in the absence of any computerized databases, may well involve TDS or a combination of TDS and middle-down sequencing (Figure 3). In Figure 3, middle-down sequencing is described with direct enzymatic generation of larger peptides that are then sequenced using TDS methods. However, as described in the past, usually there has not been a concomitant top-down determination of intact molecular weight of such proteins. Thus, unless all peptides were or are discovered and sequenced, there is no way to 100% confirm the database-suggested structures. The same problems in using bottom-up (shotgun) methods have and will then prevail using middle-down approaches. In Figure 4, middle-down is performed together with intact mass validation, overcoming the inherent disadvantages (but only if all possibly formed peptides are found and identified).
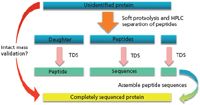
Figure 4: Middle-down protein sequencing after first doing intact mass validation (top-down proteomics) or top-down protein (TDS) on the same protein (9).
To really make middle-down the sequencing technique of choice in the future, TDS should first be done on an unidentified protein. This will provide the intact molecular ion (hence, correct molecular weight), as well as some of the N- and C-termini sequences. And then, middle-down should be performed on the intact protein, as in Figure 4, to fill in any blanks from TDS alone.
In TDS, the intact separated protein is sequenced in a variety of ways, depending on the available MS instrumentation. Still, all TDS methods and instrumental approaches in the literature today lead to significant N- and C-termini sequencing, perhaps as many as 50 residues from each end, at times encompassing all the amino acids in that protein. However, many proteins do not lend themselves to complete sequencing, and it is now impossible to predict which ones will and which ones will not before doing the needed experiments. When TDS fails, typically for very large proteins (>30 kDa), combining its results with middle-down methods, as above, can lead to a total sequencing result, including PTMs. This is the next-best result, short of having TDS alone realize the complete protein sequence and structure. What is clearly needed today, as emphasized in Krull's talk, is a better understanding of why some proteins lend themselves to TDS while many others do not. It is not clear what is involved or required to enable all proteins, regardless of size or molecular weight, to lend themselves to TDS methods. When this becomes better understood, and TDS is fully perfected, it will become possible to do sequencing of any protein, regardless of its size, molecular weight, or conformation by MS methods alone. This is but a matter of time, although how much time, nobody really knows yet. TDS is really the holy grail of protein sequencing and identifications, not just in proteomics but in all of protein analysis and characterizations.
Protein Disease Biomarkers
"Protein Disease Biomarkers: from Discovery to the Clinic" was presented by Y. Levin, currently with the Biological Services group of the Weizmann Institute of Science (Rehovot, Israel) and previously with the University of Cambridge (Cambridge, United Kingdom), and E. Schwarz and S. Bahn of the University of Cambridge. In this very interesting and potentially significant talk, proteomics was used to determine the most likely biomarkers for schizophrenia. Studies were performed at the Cambridge Center for Neuropsychiatric Research. Clinically diagnosed patients were compared to normal controls for protein differences, using nonlabeled, quantitative, bottom-up proteomics as the analytical methods. The most likely biomarkers, including several well-known proteins in healthy people, were identified as being dysregulated in diseased samples. Control studies were performed using enzyme-linked immunosorbent assay (ELISA) methods for those very proteins, involving large numbers of healthy and schizophrenic patients. Analyses of these biomarkers easily distinguished healthy from schizophrenic samples.
These analytical methods have been successfully commercialized for 51 specific protein biomarkers, and now are routinely used in clinical settings around the world, with a high success rate for diagnosing diseased states. This would appear to be one of the very best and most successful biomarker discovery projects thus far, leading to commercial kits for determining specific protein biomarkers derived from this particular disease state. Before this work, there were no biomarkers available to diagnose schizophrenia. Instead, diagnosis relied entirely on subjective and error-prone psychiatric examination and observations.
Mass Spectrometry for Protein Quality Control
Another paper very relevant to "Biotechnology Today" was also presented Monday morning. This paper, "Quality Control of Intact, Recombinant Proteins Using Sensitive, High-Resolution Mass Spectrometry," was presented by A. Ingendoh, D. Wunderlich, and C. Albers of Bruker Daltonik GmbH (Bremen, Germany). With the increased interest in newer biopharmaceuticals, proper quality control (QC) is needed to ensure use of the correct batches from the protein's production. As usual, this requires knowledge about the correct amino acid sequence, as well as characterization of PTMs. These techniques, as above, require the use of HRMS instrumentation because of the heterogeneity of most such samples, as well as the advantage provided of having an exact (!) molecular weight of the original intact protein and its enzymatic fragments. In this excellent study, the authors used an ultrahigh-resolution quadrupole time-of-flight (Q-TOF) instrument for the LC–ESI-MS analysis of various proteins, such as recombinant IgGs. These were derived from Chinese hamster ovary (CHO) cells, but a second mixture of recombinant proteins was obtained from E. coli cells. Proteins were first separated on a Zorbax SBC8 rapid resolution cartridge within 15 min, then individually analyzed by MS.
The resolution and wide mass range of this particular, Bruker-derived, instrumental setup allowed for the discrimination of discrete changes in the glycosylation patterns, even for proteins such as IgG. Further analysis of the recombinant IgG after reduction and alkylation allowed the authors to measure the MW of the light chain at 25 kDa with 0.2-ppm mass accuracy. In the second study, the MS spectra of the recombinant E. coli proteins allowed the authors to distinguish between protein monomers and dimers, covering a mass range up to 40 kDa.
The data was processed by specially designed software modules that compared mass patterns and accuracies with expected values, thereby ensuring a fast and reliable information retrieval, which is mandatory for QC. In combination with the possibility of running the intact proteins directly, a high-throughput analysis was possible. This approach lends itself immediately to semiroutine QC analyses of various batches of commercially expressed recombinant proteins as large and complicated as IgGs. It is expected that such QC methods will be, or are already, fully accepted by the regulatory agencies for lot release testing of already-approved recombinant proteins for either clinical studies before approval of a new drug application (NDA) or an abbreviated new drug application (ANDA), or after approval, when a biopharmaceutical has entered the market. It should be emphasized that other MS instrumentation vendors have also described similar analytical protocols on their commercial instrumentation, usually with equally successful outcomes for similar biopharmaceutical products.
Figure 5 illustrates some of the possible applications using HRMS for protein QC. This involved direct MS analysis of the intact IgG1, with an intact molecular weight of about 150 kDa, illustrating the presence of several variants or isoforms having different glycosylation patterns, as indicated. Using HRMS methods, it is clearly possible to identify specific glycosylation patterns for the most intense variants (bottom panel). It is also possible to visualize the entire mixture of variants for multiply charged ions, with one particular such ion at about 3043.5086 m/z. Within each such envelope resides the mixture of variants for that particular charge state (approximately 150,000/3043). This QC characterization is valuable for its ability to identify individual variants, as well as relative intensities or quantities of these, in a mixture for each batch. Such overall direct MS techniques provide rapid, reliable, accurate, precise, and absolute data to demonstrate batch-to-batch chemical equivalency or comparisons of proprietary with biogeneric samples. These latest MS techniques do not require an initial high performance liquid chromatography (HPLC) or CE separation of the variants to demonstrate their qualitative and quantitative presence. Even the lower abundance variants can be characterized in terms of their glycosylation patterns.

Figure 5: Performance of modern MS for protein QC. (Reproduced with permission of A. Ingendoh and Bruker Scientific, Bremen, Germany.)
HILIC and MS for Glycoprofiling
One final talk on Monday morning was given by P. Appelblad of Merck SeQuant AB (Umeå, Sweden), entitled "HILIC and MS — A Perfect Fit." As we have already discussed in this column, one of the requirements for sucessful characterization of recombinant glycoproteins has to do with glycan analysis, or glycoprofiling, especially for recombinant antibody products. In most instances, glycans are first released from their glycoprotein sources enzymatically or chemically, separated from the residual proteins, and then subjected to various chromatographic or electrophoretic analytical techniques. Over time, many different analytical approaches have been used, such as ion chromatography, reversed-phase chromatography, or gas chromatography. Hydrophilic interaction liquid chromatography (HILIC) is, however, the technique of choice and an attractive separation mode for polar hydrophilic compounds, such as carbohydrates. The high percentage of organic solvents, like methanol or acetonitrile, used in the mobile phases makes it the method of choice for analysis of polar compounds. HILIC is easy to use and works well where traditional reversed-phase methodology fails, and it is more intuitive and flexible than ion chromatography. The elution order is typically the opposite, with the most polar compounds eluted after the nonpolar compounds, resulting in alternative selectivity.
A bonded, zwitterionic HILIC stationary phase allows chromatographers to maximize analyte sensitivity by eluting polar compounds under a high percentage of organic mobile phase conditions and is ideally suited for MS analyte ionization and increased sensitivity for many glycan analytes. Bonded zwitterionic stationary phases are available either with silica or a polymer core-based particle, an advantage for carbohydrate analysis, as many analytes of interest are pH-dependent. The silica core solid phase has a typical working pH range of 3–8, whereas a polymeric material tolerates more acidic and more basic conditions. The use of a basic buffer component like ammonium hydroxide (NH4OH) in the mobile phase is crucial for collapsing the anomers of reducing sugars (when so desired) and aiding the separation.
Therefore, carbohydrates can be separated under both neutral conditions, and at an elevated pH, using direct ESI-MS detection. Using high-pH mobile phases, it is possible to increase mutarotation and collapse the peaks from anomers, simplifying identification of carbohydrates, even in samples with difficult matrices. Combined with simple sample preparation procedures, such as protein precipitation or liquid–liquid extraction, efficient and cost-effective analytical work schemes can be developed and used for monitoring of sugar, sugar alcohols, and other carbohydrates in different types of formulations and matrices. This then makes HILIC and MS a perfect fit for sugar analysis derived from glycoproteins, as an overall part of meeting biopharmaceutical regulatory requirements and drug substance characterizations. It is fully expected that such HILIC–MS approaches will find widespread acceptance and even more applications in the future for more and more glycoprotein biopharmaceuticals! Figure 6 (please see the print version of this issue for this figure) illustrates the LC–MS separation of various O-sialoglycopeptides at different pHs on a Merck zwitterionic ZIC-HILIC column (4).
This presentation illustrated the most recent advances in using various types of HILIC columns for a variety of carbohydrates, wherein the mobile phase conditions allowed for ideal, direct interfacing with ESI-MS. This provided for absolute glycoanalysis, even for O-sialoglycopeptides and related glycosylated species. The same approach has been effective for peptide maps (protein digests), monosaccharides, sugar alcohols, sugar phosphates, fermentation broths, neuropeptides, and metabolomics in urine samples. In most instances, more peaks were detected using HILIC than with strong cation exchange or reversed-phase methods. Multidimensional (2DLC–MS) separations using HILIC instead of strong cation exchange or reversed-phase methods in the first dimension also have led to a higher number of peaks (greater peak capacity) detected. This approach should therefore be evaluated in proteomics, peptidomics, and other -omics applications in the future. Mention should also be made of the existence today of different HILIC phases, such as ZIC-HILIC (combination of HILIC and weak ion exchange mechanisms operative), and from a variety of manufacturers and suppliers.
Quality by Design and Protein Characterization
On Tuesday morning, S. Kozlowski, Director of the Office of Biotechnology Products, OPS, CDER, FDA, presented a plenary lecture entitled "Quality by Design and Protein Characterization." Quality by design (QbD) is an initiative to improve pharmaceutical manufacturing, and it is now being applied to small-molecule development. The principles of QbD are indeed applicable to biopharmaceuticals, and there may well be some unique considerations for such products. Advances in analytical strategies are an important part of QbD implementation for complex products, and some examples of protein characterization that relate to clinical performance were described and discussed. Proteins, in general, have a large number of structural attributes, and not all attributes affect clinical performance. Kozlowski discussed potential approaches to link product attributes to safety and efficacy. QbD is a systematic approach to drug development that begins with predefined objectives and emphasizes product and process understanding and process control. It is based on sound science and quality risk management, and is described more fully in the International Conference on Harmonization (ICH) Q8R guideline on Product Development. QbD can be applied to many aspects of drug development, analytical method validation, method development, and the overall manufacturing process.
QbD involves the definition of a design space, which is the multidimensional combination and interaction of input variables (for example, material attributes) and process parameters that have been demonstrated to ensure quality. Working within the design space is not considered a change. The design space is proposed by the applicant and is subject to regulatory assessment and approval. Within the design space are the critical process parameters (CPP). These are defined as process parameters whose variability has an impact on a critical quality attribute and therefore should be monitored or controlled to ensure the process produces the desired quality (ICH Q8R).
The second, major part of Kozlowski's plenary lecture dealt with describing and defining protein heterogeneity and its various manifestations. These included PTMs such as amino acid substitution, truncation, mismatched S-S bonds, aggregation, N- and C-terminal modifications, acetylation, and numerous others. He then described the various modifications on a typical IgG molecule and where these usually occur, such as N-terminal, C-terminal, heavy chain, light chain, or glycosylation patterns and variations (Figure 7). A large amount of time was spent discussing glycosylation variants and how these affect the final, overall biological properties of the monoclonal antibody (MAb) products. The last part of this presentation dealt with immunogenicity of recombinant proteins, including how this arises, manifestations of immunogenicity in the patient, and so forth. Some discussion included pegylation of a drug substance, and how this affects shelf life, stability, half-life, pharmacokinetics, efficacy, and safety. Pegylation is a form of preformulation and has significant, positive attributes, and as a result it has become a fairly common procedure to improve the final biopharmaceutical drug product.
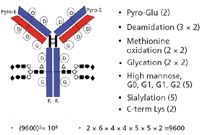
Figure 7: Attributes and combinatorics of monoclonal antibodies. (Reproduced with permission of S. Kozlowski.)
Finally, Kozlowski mentioned the incorporation of capillary electrophoresis in the biotechnology industry as a commonly used analytical method to improve overall characterization of recombinant proteins, especially antibodies. Kozlowski also mentioned its use to decipher the problems in certain commercial forms of heparin imported into the United States from China in recent years. The routine usage of modern CE by several biotech companies, such as Amgen, Genentech, and others, has now become commonplace in submittals to the FDA. Kozlowski suggested that this has been a good example of using newer modern analytical methods to improve the overall, complete, or partial structural characterization of recombinant drug substances. The related Tuesday morning session allowed for a large number of questions from the audience for both Kozlowski and Karen Russo, VP, Small Molecules, U.S. Pharmacopeia. That conversation lasted almost an hour.
Capillary Isoelectric Focusing for Monoclonal Antibody Quality and Stability Testing Although there were about two dozen poster papers presented during the two days of the meeting, only one of these was really relevant for "Biotechnology Today coverage." This was a paper entitled, "A cIEF Technique for Monoclonal Antibody Quality and Stability Testing with Biologically Functional Significance: Fragmented Antibody-CE or FAb-CESM," by M. Schwarz and F. Moffatt of Solvias AG (Basel, Switzerland). The approaches were very relevant for both intact MAbs of all types, including their aggregates, as well as to papain (or other enzymes) fragments of smaller molecular weights, including their variants. In addition, capillary isoelectric focusing (cIEF) was shown to be applicable for identification of intact variants, as in the past, but also for variants located on either the Fc or Fab portions, after papain digestion. In many instances, low levels of variants were more apparent by resolving their Fab and Fc fragments rather than the intact MAbs. Not only was cIEF ideal for studying the originally expressed intact MAb and its fragments to demonstrate lot-to-lot consistency for possible release testing, but it was also shown to be applicable for stability testing over long time frames as well as in forced degradation studies of shorter times. A series of commercial biopharmaceutical MAb products was used to illustrate the general applicability of this newer FAb-CESM method, with very similar results from antibody to antibody. Although the original studies involving cIEF to characterize MAb variants appeared over 10 years ago, its use has been widely accepted in the biotech industry for agency submittals in many countries today. This work again illustrates the general applicability and utility of cIEF but extends it now to studies involving Fab and Fc species with further advantages, as described above. Scientists in the biotech industry should take note of this work, and perhaps follow the full publications with all method details and complete results.
Acknowledgments
The authors wish to acknowledge the encouragement of Dave Walsh, former Editor of LCGC North America, in choosing the topic for this column. We also acknowledge the Chairs of the meeting, Shula Levin and Aviv Amirav, as well as Eli Grushka (Chairman of the Israel AC Society), for their help and assistance in obtaining an oral presentation for one of the authors (ISK). We thank our Israeli colleagues for their help in proofreading the final draft, vis-à-vis our describing the meeting organization itself. We also acknowledge the invaluable assistance of various colleagues who read earlier drafts of this column and made valuable suggestions for improvements (J. Klaene, S. Levin, E. Grushka, B.L. Karger, P. Appelblad, F. Moffatt, and others). All responsibility for discussions and interpretations of the oral presentations or poster papers, as discussed above, remains that of the column editors (ISK and AR). Finally, we wish to acknowledge the excellent administrative expertise provided by BioForum, Ltd., in organizing, managing, running, and overseeing the entire meeting, so that it realized the highest level of professional success.
Editor note: Figures 1, 2, and 6 were only available for use in the June print issue. If you would like a copy of this issue please contact Managing Editor, Meg Evans (mevans@advanstar.com) and a copy will be mailed to you.
Simion Kreimer is a graduate student at Northeastern University pursuing a master's degree in the field of Biopharmaceutical Regulatory Sciences. He has worked as a technician for MSM Protein Technologies, a company specializing in monoclonal therapeutic antibody discovery. His research interests include MS application in protein characterization and proteomic research for the purpose of metabolic engineering.

Simion Kreimer
Ira S. Krull is Professor Emeritus of Chemistry and Chemical Biology at Northeastern University, Boston, Massachusetts, and a member of LCGC's editorial advisory board.

Ira S. Krull
Anurag S. Rathore is a biotech CMC consultant and an associate professor with the Department of Chemical Engineering, Indian Institute of Delhi, India.

Anurag S. Rathore
References
(1) S. Wu, H. Jiang, W. Hancock, and B.L. Karger, Anal. Chem. 82(12), 5296 (2010).
(2) H. Jiang, S. Wu, B.L. Karger, and W. Hancock, Anal. Chem. 82(14), 6154 (2010).
(3) I.S. Krull and A. Rathore. LCGC North America 28(12), 1028 (2010).
(4) O. Hernandez-Hernandez, R. Lebron-Aguilar, J.E. Quintanilla-Lopez, M. Luz Sanz, and F.J. Moreno, Proteomics 10, 3699 (2010).

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)









