The Use of Light-Scattering Detection with SEC and HPLC for Protein and Antibody Studies, Part I: Background, Theory, and Potential Uses
LCGC North America
Light-scattering detection: the theory and the advantages
Light-scattering detection, particularly combined with size-exclusion chromatography or high performance liquid chromatography, is very effective for the characterization of proteins and antibodies. Examples of applications include determining absolute molecular weight, studying PEGylated proteins, characterizing protein–drug and antibody–drug conjugates, and studying protein aggregation.
Light-scattering (LS) detection, particularly multiple-angle light scattering (MALS) can be very useful for the characterization of proteins and antibodies. In a number of applications, its performance exceeds that of mass spectrometry (MS) detection. Here in part I of this two-part column, we outline the development of light scattering in biotechnology, summarize its current uses and advantages, and explain the theory of how light scattering works. In part II, we provide a few detailed examples of its use and discuss how its performance compares to MS and other methods.
History of Light Scattering
In 1974, Ouano and Kaye ushered in a new era for macromolecular characterization with the first use of on-line light-scattering detection (1,2).
Despite the intrinsic appeal of this concept, however, many improvements in both the chromatography and detection were required for further implementation with relative ease. Important developments included the detection and quantitation of branching, precise measurement of the polydispersity of so-called "standards," conformation plots, precise extraction of M-H-S coefficients by combining light-scattering and viscosity measurements with reversed-phase chromatography to detect and quantify multimeric formation, field-flow fractionation (FFF) for an alternative separation combined with light scattering, temperature rising elution fractionation (TREF) measurements, capillary electrophoresis (CE) combined with light scattering, and the quantitation of aggregation phenomena and microgel formation.
Light Scattering in the Study of Biopolymers
In the mid-1980s there were few, if any, applications of chromatography combined with any form of light-scattering detection for the study of biopharmaceuticals described in the literature (3–6). This was true even for conventionally derived proteins from natural sources (that is, not recombinant proteins). Most detectors used in tandem with size-exclusion chromatography (SEC) or high performance liquid chromatography chromatography (HPLC) for biopolymers were ultraviolet (UV), differential refractive index (DRI), photodiode array (PDA), ultraviolet–visible (UV–vis), or fluorescent. Back then, MS was not yet conveniently interfaced with any form of SEC or HPLC. There was no general usage of electrospray ionization (ESI) or matrix-assisted laser desorption–ionization-time of flight (MALDI-TOF) MS developed or applied for biopolymers. Most vendors of light-scattering instrumentation had not yet developed instrumentation or applications aimed at the nascent, but rapidly growing biopharmaceutical industry. This situation would rapidly change.
Our own light-scattering work (Krull and colleagues) in those days was supported and encouraged by a firm known as Laboratory Data Control/Milton Roy (LDC/MR), in Florida, which no longer exists. This company was involved in both HPLC and light-scattering instrumentation, but it manufactured only a low-angle laser light scattering (LALLS) instrument and an off-line, differential refractometer for measuring the differential or incremental refractive index. Both instruments used a 633-nm laser. Of course, LALLS had been used for synthetic organic polymer characterizations but there very few, if any, applications in the literature for using LALLS or other light scattering, with biopolymers separated using HPLC or SEC (3–16) In our initial discussions with the management of LDC/MR, we proposed to study such new and novel applications, and to bring LALLS/DRI into a more common usage for biopharmaceutical proteins, antibodies, and related analytes. Herb Kenny and colleagues were influential in bringing about this collaboration, which lasted several years. Eventually LDC/MR was purchased by Thermo Electron Corporation, and the light-scattering product line was subsequently abandoned. Other vendors picked up the slack of course, and today there are at least two major companies offering such instrumentation with numerous applications for biopharmaceuticals: Wyatt Technology and Malvern Instruments. There are some other, smaller firms as well, but these do not offer the same extent of instrumentation diversity, in-house applications literature, or technical expertise.
In the late 1980s and early 1990s, Krull and others used SEC or HPLC with isocratic elution conditions interfaced on-line and in real-time with LALLS, using off-line DRI for dn/dc measurements of the protein analytes. Later, we moved to using either isocratic or gradient elution in ion-exchange chromatography (IEC), hydrophobic interaction chromatography (HIC), or reversed-phase chromatography (3–16). We also began using on-line, 633-nm DRI with UV detection (concentration detector) to obtain at least three chromatograms (light scattering, UV, and DRI). Such approaches then provided direct light-scattering, dn/dc, and c (concentration) measurements on every injection of protein products and mixtures. With this approach, we could derive very accurate and precise measurements of molecular weights (weight-average, z-average, and n-average) but we could not yet calculate the radius of gyration (Rg)or the hydrodynamic radius (Rh). We could also obtain the second virial coefficient term (A2) in the Zimm equation, suggesting the best solvents to use for nonaggregation or disaggregation prevention and overall stability or compatibility of the proteins with the HPLC solvents for their elution and separations. Eventually, we were able to use viscometry instrumentation, in an on-line format with the light-scattering, UV, and DRI detectors, and then derive Rh in addition to the measurements previously described. These were studies in the late 1980s and early 1990s, using instruments that are by and large no longer commercially available. However, the techniques were soon readily adopted and adapted by various biotechnology firms for their own recombinant protein or antibody products, perhaps using other commercial instrumentation and methods, but especially SEC–MALS (17).
Current Uses of Light Scattering in the Biotechnology Industry
The use of SEC–MALS and similar techniques is still growing within the biotechnology industry. The most common uses are outlined below.
Determining Absolute Molecular Weight
In general, today most applications of SEC–LS involve the determination of absolute molecular weights, as described above, plus either Rh or Rg. By absolute, we mean without reference to standards whose molecular weights are often known and used to calibrate the instrumentation. Nor does absolute refer to any technique that makes use of calibration curves such as SEC or intrinsic viscosity based on so-called "universal calibration."
That is, light scattering can be used to determine absolute molecular weights ±2–3% (standard deviation) or better for individual proteins after HPLC or SEC separations, and for covalent (oligomers) or noncovalent aggregates of such proteins with one another or with their antibodies. As long as the conditions being used in HPLC or SEC do not change the nature of the protein samples through the chromatographic process, then light-scattering analysis will provide a true picture of what was in the injected sample in terms of monomer, dimer, or higher-order aggregates of the parent protein or antibody.
Of the four absolute methods for measuring molecular weights, light scattering is applicable to the greatest range of samples. The other three methods capable of absolute molecular weight determination are sedimentation equilibrium, vapor osmometry, and MS (although the latter method is not as useful if the "solution" properties of the molecules, such as noncovalent aggregates, are being measured). For light scattering, the range of accessible molecular weights is from several hundred to tens of millions of daltons (grams per mole). Vapor osmometry measurements span a much smaller range, up to only a few hundred thousand daltons under the most favorable of conditions. The lower limit, however, can be an order of magnitude smaller. Sedimentation equilibrium, also called analytical ultracentrifugation, often requires several days of measurement and has a range of applicability considerably smaller than light scattering. MS has an upper limit between 500,000 and 1,000,000 Da, although today, it can be considerably higher, in the tens of millions, with special instrumentation.
Light-scattering techniques also have another interesting advantage when combined with fractionation techniques such as SEC: They can be used to determine the molecular weight distribution and various other molecular properties, as we shall see in the following sections and in part II.
Studying PEGylated Proteins
PEGylation (adding a molecule of polyethylene glycol [PEG]) is a widely used technique for modifying protein drugs to make them more water soluble and provide different pharmacokinetic properties. HPLC–LS or SEC–LS can be used to determine the nature of PEGylated proteins, or of proteins that have been PEGylated and perhaps then modified with another, synthetic organic polymeric reagent (17,18). It is now possible to determine the molecular weight ratio of protein to PEG in a PEGylated protein. This is also true for virtually any polymer used to modify a protein by covalent or even noncovalent steps. Electron-transfer dissociation (ETD) in MS has also proven useful for identifying the specific site and nature of PEGylation in certain peptides (19). However, this technique does not work when trying to determine the molar ratio of protein to PEG for a mixture of PEGylated proteins.
Characterizing Drug Conjugates
Another very important and growing area of biotechnology characterization relates to antibody–drug and protein–drug conjugates, which are synthetically derived combinations of an antibody or protein and a small-molecule drug against a specific disease related to that protein target. Using LS-based techniques, it is possible to measure, with a high degree of accuracy, how much drug (or PEGylation) has been incorporated, on the average of all such species present. It cannot, however, indicate the specific points of such attachments. On the other hand, if these derivatives can be adequately resolved before MS detection, MS can indicate exactly where a drug or PEG molecule is located on a (specific) protein sequence; however, MS cannot accurately measure how much of the drug has been incorporated into all the protein molecules unless each such species can be resolved one from the next. This is yet another area where the capabilities of light scattering are superior to those of MS.
Studying Aggregation
Perhaps the most useful information for the biotech industry is that which indicates the level of aggregation present in any protein or antibody sample, as well as the specific nature of each aggregate (dimer, trimer, and so on), and the Rh or Rg of each aggregate. Another piece of information of value will be a demonstration that the aggregates being measured by SEC with LS detection were indeed present in the original sample, and that their amounts (relative percent peak areas) have not changed through the chromatographic and detection steps.
Light scattering by itself does not change the nature of aggregation in any given sample, but that may not always be true for SEC and HPLC or other modes of separations, or when using MS detection methods. At times, SEC and MS have both been shown to change the nature of aggregates detected after initial injection into a separation-MS scheme (20–22). Furthermore, the level of aggregates could be made to increase or decrease depending on the conditions used in both the separation (SEC) and MS steps.
This, of course, is not a desirable occurrence. If an analytical method changes the original sample components, then it becomes useless in determining what was originally in the sample before analysis. Analysts must demonstrate using accepted methods that their analytical methods have not in any way altered the original samples through the analytical scheme. If this cannot be done, then one should assume the worst: that there are indefinable alterations of the sample components during that analytical method. In essence, that makes all data and conclusions questionable at best and, in reality, worthless.
Even gel permeation chromatography (GPC) and SEC are known to change sample composition, especially in the case of noncovalent aggregation. (In the case of covalent aggregates, other than for disulfide rearrangements, there are usually no changes in the sample composition by HPLC or ultrahigh-pressure liquid chromatography [UHPLC] methods.) In the case of a complex mixture of synthetic, organic, or natural polymers, changes in elution times because of the particular GPC or SEC conditions being used may suggest differing molecular weights as a function of concentrations injected, which is artifactual. This concentration dependence will mainly occur when using a reference calibration method but should not occur when using an absolute technique such as light scattering. Of course, if some of the injected biopolymer mixture remains on the SEC column, then light scattering will provide the true molecular weight for what is being eluted but not necessarily what was in the sample injected. Knowing when this is happening is, of course, extremely important in characterizing the original sample.
In part II of this column series, we provide further discussion of some of the possible pitfalls in attempting to characterize noncovalent protein aggregates using today's MS methods. Such approaches can be fraught with danger, unless the analyst has very clearly and definitively demonstrated that his or her methods do not and will not alter the sample components or their relative amounts.
Today, most applications of light scattering in the biopharmaceutical industry are done using MALS–RI–UV combinations, or some other type of instrumentation arrangement (such as RALS–LALS).
Light-Scattering Theory
Before we enter into further detail (in part II) about the application of light-scattering methods to the analysis of biopharmaceuticals, let us review how light scattering works.
Classical light-scattering measurements, in the absence of initial separations (such as SEC), are performed by first preparing a series of samples at different, decreasing concentrations in a suitable solvent. The solvent chosen must be fully compatible with the analytes and should not in any way change the nature of the original sample. Each sample is then illuminated sequentially by a collimated light source, such as a monochromatic laser, and the scattered light from each sample is measured as a function of scattering angle (1,2,23–29). This is shown schematically in Figure 1, where an array of collimated detector elements detect scattered light at discrete scattering angles (30).

Figure 1: An array of collimated detector elements used to detect scattered light at discrete scattering angles.
Historically, such measurements were first made using a single detector that rotated about the sample cell, stopping at each angular location, where measurements were made. Using an array of detectors speeds up the measurement process significantly. The relation between the measured intensities and the weight-average molecular weight (MW) is given to the second order in the sample concentration (c), by the equation developed by Zimm:
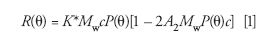
Zimm showed that for such small concentrations, equation 1 may be written in its reciprocal form as
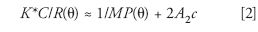
In equations 1 and 2,
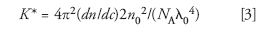
where c is the concentration of the solute molecules (g/mL), n0 is the refractive index of the solvent, and dn/dc is the refractive index increment of the solution (that is, the solution refractive index changes an amount dn for a solute concentration change of dc). Furthermore,
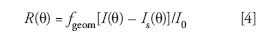
where, θ is the scattering angle, I(θ) is the intensity scattered by the solution into the collimated detector solid angle aboutθ, Is (θ) is the intensity scattered by the solvent into the collimated detector solid angle about θ, and I0 the light intensity incident on the sample. fgeom is a geometrical factor depending on the structure of the scattering cell, the refractive indices of the solvent and the cell, as well as the field of view of the corresponding detector and its acceptance angle for scattered light. NA is Avogadro's number, λ0 is the incident wavelength in vacuum, MW is the weight-average molecular weight, and A2 is the second virial coefficient (a measure of the solvent-solute interaction). The form factor, P(θ), may always be written as an infinite series in sin2θ/2:
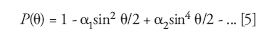
where α1 and α2 are constants and
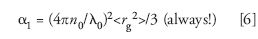
and
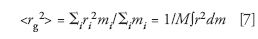
the integration is taken over all mass elements, dm, of the molecule. The distance of mass element dm from the molecule's center of gravity is r.
Measurements of the scattered light intensity from different sample concentrations form the basis of Zimm's method to extract the molecular parameters Mw, <rg2>, and A2. Further details may be found in the original Zimm papers (26,27,33). Note from equation 2 above, in the limit, as c and the scattering angle, e, go to zero, K*c/R(0°) = 1/Mw. Although the molecular weight derived in this manner is the Mw of the sample, the derived mean square radius <rg2> is, in general, some type of average that is only easily defined for the case of random coil molecules in a theta solvent (A2 = 0). In that case, it may be shown quite easily that the measured mean square radius is a z-average value.
Combining Light Scattering with Chromatography for Full Characterization
Although light-scattering measurements performed on unfractionated samples will yield average values for both mass and size, following Zimm's method (discussed briefly in the previous section), the true characterization of any nonmonodisperse sample requires that the differential and cumulative distributions be measured. To achieve this, the sample must first be fractionated, for instance by SEC or GPC. Figure 2 shows a typical SEC arrangement combined with a light-scattering detector.

Figure 2: A typical SEC arrangement combining UV, MALS, and RI detection in series.
The light-scattering detector is usually placed between the concentration detector (a DRI or UV detector) and the SEC columns (31). Samples separated by the columns are generally diluted between 10 and 100 times from the injected concentration (which, in itself, can often lead to varying degrees of disaggregation, if aggregates were first present in the injected sample), so the second virial coefficient term in equation 2 may generally be neglected. Therefore, for GPC or SEC, equations 1 and 2 both reduce to the form
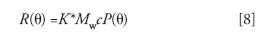
It is important to note that the baseline adjusted light-scattering signal (excess Rayleigh ratio) is proportional to the product of the molecular weight times c. Because c is proportional to the number density multiplied by the molecular weight, the excess Rayleigh ratio is proportional to the number of molecules per unit volume multiplied by the square of the molecular weight. Thus, the excess Rayleigh ratio, which is measured at each eluted fraction (slice), may be extrapolated to zero scattering angle to determine the molecular weight for that fraction from the intercept with the ordinate axis.
Because the fractionation process itself is assumed to yield monodisperse fractions at each collection, the weight-, number-, and z-average molecular weights should be the same at each such slice. The concentration (UV or RI) detector measures a concentration at each slice; therefore, one can calculate the differential and cumulative-weight fraction molecular weight for each sample injected. The mean square radius may be calculated at each slice from the slope of the excess Rayleigh ratio as a function of sin2θ/2 extrapolated to zero scattering angle, and thus the distributions of the mean square radius may also be calculated. Note that the mean square radius may be calculated without any knowledge of the sample concentration, provided that the result of equation 8 holds.
From a measurement of the root mean square radius and the corresponding mass at each slice, the conformation of the molecules making up the distribution present in the sample ensemble may be determined, as well. There are two requirements for this to be possible: First, the molecules must be large enough to permit a meaningful measurement of the mean square radius and, second, the distribution present must be polydisperse and span a reasonable range of molecular weights. Finally, it should be noted that for certain types of copolymers, the mean square radius cannot be calculated immediately from the variation of the excess Rayleigh ratio with sin2θ/2.
Conclusion
Here in part I of this two-part column, we have discussed the history of light-scattering detection in tandem with chromatography for biopolymer separations and characterizations, and briefly discussed current applications. We have also provided a summary of the theory of light scattering and how it is combined with chromatography. In part II, we will provide a more in-depth discussion of several applications of light scattering in biotechnology: measuring protein stability, measuring aggregates in formulation studies, and characterizing low-molecular-weight heparin. We also compare the performance of light-scattering methods with MS detection and other methods.
Acknowledgments
This column has been a group effort, involving several unsung contributors, as well as those whose names appear on the first page. More specifically, we are indebted to several colleagues at Wyatt Technology in Santa Barbara, California, who read various sections, always making constructive and useful, suggested revisions. (Often, we actually listened to such suggestions.) In particular, our acknowledgement and appreciation goes to Phil Wyatt for the description of the fundamentals of light-scattering theory and equations and to John Champagne for the section on using light scattering without chromatography in free solution. Other people we wish to ackowledge at Wyatt include Cliff Wyatt, Geof Wyatt, Michelle Chen, and Sigrid Kuebler. Any errors of omission or commission are clearly those of the editors and co-author (S.K.) alone.
References
(1) Wikipedia, en.wikipedia.org/wiki/Multiangle_light_scattering.
(2) Laser Light Scattering: www.ap-lab.com/light_scattering.htm.
(3) I.S. Krull, H. Stuting, and S. Kryzsko, J. Chromatogr. 442, 29 (1988).
(4) I.S. Krull, R. Mhatre, and H.H. Stuting, Trends in Anal. Chem. 8(7), 260 (1989).
(5) H.H. Stuting, I.S. Krull, R. Mhatre, S. Krzysko, and H. Barth, LCGC North Amer. 7(5), 402 (1989).
(6) R. Mhatre, H.H. Stuting, and I.S. Krull, J. Chromatogr. 502(1), 21 (1990).
(7) H.H. Stuting and I.S. Krull, Anal. Chem. 62, 2107 (1990).
(8) R. Mhatre and I.S. Krull, J. Chromatogr. 591, 139 (1992).
(9) R. Mhatre and I.S. Krull, Chromatographia 34, 357 (1992).
(10) D.J. Mageira and I.S. Krull, J. Chromatogr. 606, 264 (1992).
(11) R. Mhatre and I.S. Krull, Anal. Chem. 65, 283 (1993).
(12) R. Mhatre, R.-L. Qian, I.S. Krull, S. Gadam, and S. Cramer, Chromatographia 38(5/6), 349 (1994).
(13) I.S. Krull, R. Mhatre, and J. Cunniff, LCGC North Amer. 13, 30 (1995).
(14) M.E. Szulc, R. Mhatre, J. Mazzeo, and I.S. Krull, in High Resolution Separation of Biological Macromolecules, Methods in Enzymology Series, B.L. Karger and W. Hancock, Eds. (Academic Press, Waltham, Massachusetts, 1996), Chapter 8, p. 175.
(15) R.-L. Qian, R. Mhatre, and I.S. Krull, J. Chromatogr. A 787, 101 (1997).
(16) L.C. Santora, P. Sakorafas, Z. Kaymakcalan, I.S. Krull, and K. Grant, Anal. Biochem. 299(2), 119 (2001).
(17) Protein Pegylation Application Note, Protein Solutions, Wyatt Technology, Santa Barbara, California, 1996.
(18) Pegylated Protein Modifications Application Notes, Protein Solutions, Wyatt Technology, Santa Barbara, California, 1997.
(19) T.P. Second, A.W. Carr, R.C. Cummins, R. Viner, and L. Huang, "Characterization of PEGylated Peptides and Site Localization of Attachment with High Resolution ETD Mass Spectrometry," Thermo Fisher Scientific, Fremont, California, Application Note (2010).
(20) I.S. Krull, R. Mhatre, and J. Cunniff, LCGC North Amer. 12(12), 914 (1994).
(21) M.E. Szulc and I.S. Krull, in Practical HPLC Method Development, Second Edition, L. Snyder, J. Glajch, and J.J. Kirkland, Eds. (J. Wiley & Sons, Inc., New York, New York, 1997), Chapter 3.
(22) M.E. Szulc, R. Mhatre, J. Mazzeo, and I.S. Krull, in High Resolution Separation of Biological Macromolecules, Methods in Enzymology Series, B.L. Karger and W. Hancock, Eds. (Academic Press, Waltham, Massachusetts, 1996), Chapter 8, p. 175.
(23) C. Laue and D. Hunkeler, Int. J. Polym. Anal. Charact. 5, 511–529 (2000).
(24) T. Wang and J.A. Lucey, J. Dairy Sci. 86(10), 3090–3101 (2003).
(25) A. Oliva, M. Llabres, and J.B. Farina, Curr. Drug Discov. Technol. 1(3), 229–242 (2004).
(26) B.H. Zimm, J. Chem. Phys. 16, 1099–1116 (1948).
(27) P.J. Wyatt, Appl. Opt. 7(10), 1879–1896 (1968).
(28) W. Kaye and J.B. McDaniel, Appl. Opt. 13(8), 1934–1937 (1974).
(29) W. Schartl, Light Scattering from Polymer Solutions and Nanoparticle Dispersions (Springer-Verlag, Berlin, Germany, 2010).
(30) Absolute Characterization of Proteins and Biopolymers: Combining SEC with Light Scattering Detection, S.C. Kuebler, Wyatt Corporation Webinar, online at www.wyatt.com, Figure 7.
(31) Absolute Characterization of Proteins and Biopolymers: Combining SEC with Light Scattering Detection, S.C. Kuebler, Wyatt Corporation Webinar, online at www.wyatt.com, Figure 12.
(32) B.H. Zimm, J. Chem. Phys. 13(4), 141 (1945).
Ira S. Krull is Professor Emeritus of Chemistry and Chemical Biology at Northeastern University, Boston, Massachusetts, and a member of LCGC's editorial advisory board.

Ira S. Krull
Simion Kreimer is a graduate student at Northeastern University pursuing a doctorate in the fields of chemistry and chemical biology through research in the Barnett Institute. His research interests include the application of MS and chromatography to translational medicine and pharmacology.

Simion Kreimer
Anurag S. Rathore is a biotech CMC consultant and an associate professor with the Department of Chemical Engineering at the Indian Institute of Delhi, India.

Anurag S. Rathore












