Online HPLC–HRMS Platform: The Next-Generation Process Analytical Technology Tool for Real-Time Monitoring of Antibody Quality Attributes in Biopharmaceutical Processes
Online monitoring of critical quality attributes (CQAs) directly within the bioreactor can provide the basis for advanced processing of therapeutics production, including automated real-time monitoring, feedback control process intensification, smart manufacturing, and real-time release testing. This paper presents recent developments in online high performance liquid chromatography–high-resolution mass spectrometry (HPLC–HRMS) platforms as a promising process analytical technology (PAT) tool for real-time monitoring of antibody quality attributes (QAs) in biopharmaceutical processes. This technology can be used to monitor multiple CQAs and process parameters during cell culture production, enabling real-time decisions.
In 2021, the U.S. Food and Drug Administration (FDA) agency published a guidance document for the biopharmaceutical industry titled “Q13 Continuous Manufacturing of Drug Substances and Drug Products” (1). In this guidance, the FDA specifically supports the development and implementation of innovative online analytical technologies that can improve drug quality, enhance efficacy, and boost speed to market. Recently, there have been increased efforts to develop new process analytical technology (PAT) tools applied to real-time monitoring of cell culture processes directly within the bioreactor (2). Many of these PAT tools being in the form of sensors or spectroscopic probes, such as pH, temperature, near-infrared (NIR), mid-infrared (MIR), and Raman spectroscopy.
Most efforts focused on improving the means of monitoring cell culture process performance and parameters, such as (but not limited to) online glucose concentration via Raman spectroscopy (3) and pH via drift-free pH probes (4), but only a few PAT tools can directly measure the product quality attributes (QAs) (including oxidation, deamidation, and glycosylation). This is the result of several reasons, such as (i) few commercially available tools for online monitoring of critical quality attributes (CQAs); (ii) low concentration of the attributes, thus requiring specialized and non-automatable instrumentation; and (iii) significant burden to demonstrate comparability with traditional offline assays. Some studies reported the use of chemometric models to correlate product QAs with spectroscopic signals, thus indirectly measuring the trending of CQAs during the cell culture production process (5). However, these models are often product-specific, and typically require large data sets to validate the chemometric models. In addition, when the manufacturing process conditions change, which is quite common during process development, the model must be re-optimized or re-validated. High performance liquid chromatography (HPLC) has been the main workhorse in the biopharmaceutical industry, from research and development (R&D) to quality control (QC) laboratories (6). The utility of HPLC has been further enhanced with the recent emergence of multi-attribute methods (MAM) by combining, for example, HPLC with high-resolution mass spectrometry (HRMS). The peptide mapping-based MAM approach has the potential to replace conventional methods (such as ion exchange for charge variants analysis, reduced capillary gel electrophoresis for polypeptide cleavage variants analysis, or hydrophilic interaction chromatography for glycan profiling) for product release, because of its capacity to simultaneously monitor several CQAs (7). However, the application of MAM, focusing on in-process samples analysis (during scale-up and technology transfer), has been lacking.
Nonetheless, leading bio- pharmaceutical companies have recently published interesting applications that integrate HPLC into production processes for CQAs monitoring with HRMS detection. Liu and associates (8) demonstrated the success- ful application of a HPLC-tandem mass spectrometry (MS/MS)-based MAM method to monitor multiple monoclonal antibody product QAs throughout the cell culture process. A comparison between two different scales, a 3 L bioreactor and a 2000 L single use bioreactor in the good manufacturing practice (GMP) pilot plant, using daily in-process samples, confirmed the overall comparable QA profile of the final product.
Other applications have also been reported describing how time-consuming manual peptide mapping sample preparation steps can be replaced with automated workflows (9). For example, a Protein-A column used for antibody purification was combined with an automated peptide mapping (such as reduction, digestion, and separation of peptides) workflow (10). The method enabled monitoring of common post-translational modifications (PTMs) such as oxidation, deamidation, and succinimide formation from harvest cell culture fluid at different purification or production steps. For instance, the deamidation is known to reduce potency in some biotherapeutic products (11). This proof-of-concept study demonstrates how this approach can be used to inform the process and make timely adjustments, including (i) adjusting the temperature of the cell culture; (ii) reducing the pH of the cell culture; (iii) adjusting the level of carbonate in the cell culture; or (iv) stopping the cell culture and harvesting the protein. Therefore, rather than discarding cell culture batches that do not meet the quality specifications following offline analysis, the process can be adjusted, avoiding significant burden on the manufacturing operations.
Automated cell culture sampling systems were recently developed by various manufacturers, making it possible to monitor product quality attributes directly from the bioreactors. These systems, which allow samples to be taken at predefined intervals or scheduled times, are capable of removing cell-free material from the bioreactor and performing automated sample preparation prior to HPLC analysis in a fully automated manner. The HPLC analysis can be also preprogrammed via a vendor-provided driver to operate over an entire cell culture production process without manual operations. By doing so, the operator can generate significant data to allow robust process control. As an example, a fully integrated online HPLC–MS system including modular automated sampling technology (MAST)-based aseptic sampling, multi-function sequential injection analysis (SIA) sample preparation, and HPLC–HRMS analysis has been developed for an automatic in-process monitoring of multiple QAs of a model mAb over a 17-day cell culture process (see Figure 1) (12).
FIGURE 1: Schematic representation of the fully automated HPLC–HRMS platform for the real-time monitoring of antibody QAs for cell culture processes in bioreactors. The system is composed of online sampling liquid from the bioreactor, chromatographic purification of protein from the sampled liquid (thereby providing purified protein), and analysis of the purified protein by HPLC–HRMS.
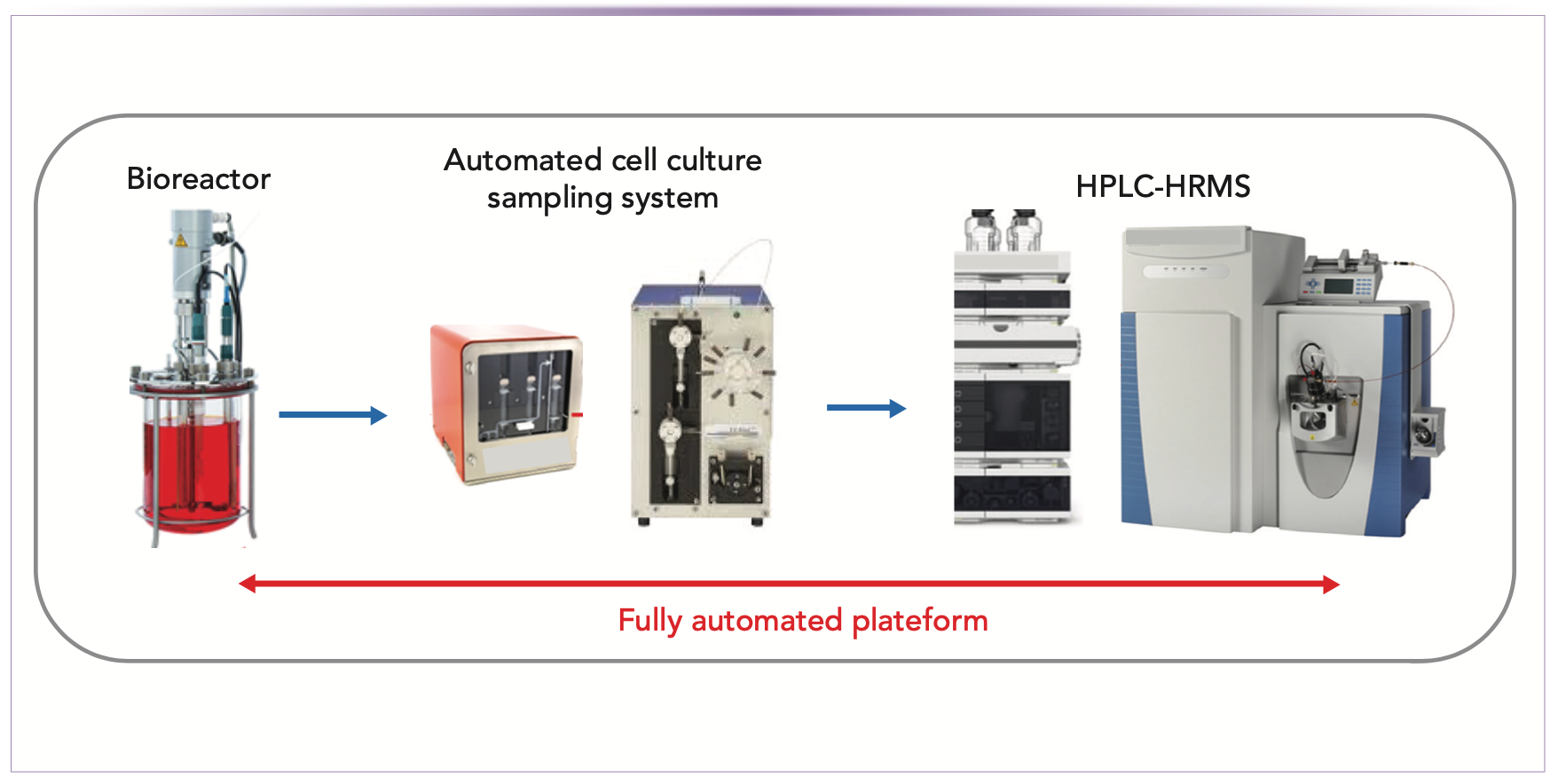
This study demonstrates that online HPLC–HRMS analytical workflows offer the possibility to modulate cell culture process conditions, resulting in a product QA profile within the quality specifications. The high-flexibility of these online systems enables additional QAs, such as titer, charge, or size variants distribution, to be monitored. The automated monitoring of titer can also be used for the generation of large data sets to help with building predictive models for spectroscopic analysis, such as Raman spectroscopy.
Overall, the demonstrated capability of these systems to monitor multiple QAs at both the intact and peptide level on the same platform, and without manual intervention, provides a rapid and reliable characterization, while enabling feedback control schemes and enhanced data generation for process development.
As such, online HPLC–MS platforms have advanced processing modes, including real-time monitoring and feedback control aimed at improving the process yield as well as the product quality. In the near future, replacing the sophisticated HRMS with low-cost simple detectors would be an alternative to reduce the barrier for the broad adoption of online platforms, especially in a GMP environment, for automated feedback control of bioprocesses, and for routine in-process CQA monitoring.
References
(1) US Food and Drug Administration, Guidance for Industry: Continuous Manufacturing of Drug Substances and Drug Products (U.S. Department of Health and Human Services, Food and Drug Administration, Rockville, MD, 2021).
(2) E.J. Kim, J.H. Kim, M.-S. Kim, S.H. Jeong, and D.H. Choi, Pharmaceutics 13(6), 919 (2021).
(3) B. Kozma, E. Hirsch, S. Gergely, L. Párta, H. Pataki, and A. Salgó, J. Pharm. Biomed. 145, 346–355 (2017).
(4) V. Saucedo, B. Wolk, A. Arroyo, and C.D. Feng, Biotechnol. Progress 27(3), 885–890 (2011).
(5) K. Buckley and A.G. Ryder, Appl. Spectrosc. 71(6), 1085–1116 (2017).
(6) Handbook of Pharmaceutical Analysis by HPLC (Elsevier Academic Press, Amsterdam, The Netherlands, 2005).
(7) R.S. Rogers, M. Abernathy, D.D. Richardson, J.C. Roue, J.B. Sperry, P. Swann, et al, AAPS J. 20(1), 7 (2018).
(8) Y. Liu, J. Fernandez, Z. Pu, H. Zhang, L. Cao, I. Aguilar, et al, J. Pharm. Sci. 109(11), 3319–3329 (2020).
(9) J. Camperi, A. Goyon, D. Guillarme, K. Zhang, and C. Stella, Analyst 146, 747–769 (2021). doi:10.1039/ D0AN01963A
(10) J. Camperi, L. Dai, D. Guillarme, and C. Stella, Anal. Chem. 92(12), 8506–8513 (2020). doi:10.1021/acs.analchem.0c01250
(11) R.J. Harris, B. Kabakoff, F.D. Macchi, F.J. Shen, M. Kwong, J.D. Andya, et al, J. Chromatogr. B Biomed. Appl. 752(2), 233–245 (2001).
(12) Y. Liu, C. Zhang, J. Chen, J. Fernandez, P. Vellala, T.A. Kulkarni, et al, J. Pharm. Sci. 111(2), 358–367 (2022). doi:10.1016/j.xphs.2021.09.011
Julien Camperi is a Technical Development Scientist with the Cell Therapy Engineering & Development department of Genentech in South San Francisco, California. Direct correspondence to: camperi.julien@gene.com.

Accelerating Monoclonal Antibody Quality Control: The Role of LC–MS in Upstream Bioprocessing
This study highlights the promising potential of LC–MS as a powerful tool for mAb quality control within the context of upstream processing.
Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)













