New Frontiers for Mass Spectrometry in Lipidomics (Part 1)
LCGC Europe
This two-part column explains how lipids analysis has evolved and the variety of techniques and approaches currently used in lipidomics, including their advantages and disadvantages, such as sample preparation, separations and, of course, mass spectrometry.
The study of lipid biology is undergoing a remarkable, technology-driven transformation that most notably involves mass spectrometry (MS) and its ancillary techniques, such as liquid chromatography (LC) and ionization sources. Greater investment inflows and a surge of activity, especially in the field of drug and biomarker discovery, is fuelling the growth of global lipid analysis — lipidomics — making it a standard research tool in academic, pharmaceutical and biotechnology sectors. Additional areas of interest in lipidomics include plant, microbial and nutritional research.
Why Lipids?
Lipids are absolutely essential for life, playing diverse and important roles in nutrition and health. Alterations in lipid metabolites are associated with various human diseases including obesity, heart disease and diabetes mellitus (1,2). The ability to profile the lipid composition of biological samples is important in disease diagnosis and drug discovery, attracting strong social and economic interests. For example, the discovery that cholesterol and triglycerides are linked to heart disease affected clinical testing, as well as drug, food and lifestyle enterprises. Indeed, for the last 50 years, we experienced a lipid phobia, the emblematic manifestations of which are readily apparent in the many food labels advertising "low fat" and "low cholesterol". Cholesterol-lowering agents, such as statins like atorvastatin (Lipitor, from Pfizer, New York City, New York, USA), are among the most successful drug classes, accounting for $21.5 billion a year market in the United States. Other commonplace drugs, such as nonsteroidal anti-inflammatory drugs including acetylsalicylic acid (aspirin) and celecoxib (Celebrex; Pfizer), are also directed against lipidmetabolizing enzymes. However, not all lipids are bad for us. Actually, some lipids promote health, such as omega-3 fatty acids and vitamins A, D and E, which explains their exponential growth as food supplements and nutraceuticals.
New research in lipid biology suggests that, especially in the areas of drug discovery and disease diagnostics, we may have formerly adopted a myopic view of lipid metabolism. For years, biomedical research focused on analysing only a handful of lipids. From the extrapolated results of those narrowly focused analyses, we sought to understand more than we could reasonably expect to know: the multifaceted mechanisms of action and the effects of drugs, the causes of complex diseases and the intricate biological alterations associated with those diseases.
As we move into a new era of lipid analysis, the potential to accurately and rapidly measure hundreds of individual molecular species provides the opportunity to use more complex lipid profiles for drug discovery and disease diagnostics. Because lipids are present in all living organisms, other areas of applications such as plant, microbial and nutritional research could also benefit from improvements in lipid analysis and a better understanding of lipid metabolism.
What is a Lipid?
Lipids constitute one of the largest classes of biological macromolecules (Figure 1). Together with nucleic acids, proteins and carbohydrates, they are present in living organisms that span the spectrum of biological complexity: animals, plants, fungi, protists, bacteria, archaea and viruses. Chemically, lipids are hydrophobic or amphipathic small molecules (<1500 Da) of biosynthetic origin, which can be counted on the order of tens of thousands. Lipids have enormously diverse chemical structures and are classified into eight main categories (3):
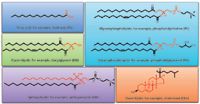
Figure 1: Representative structures for major lipid categories and examples of core structures in red.
- Fatty acyl
- Glycerolipids
- Glycerophospholipids
- Sphingolipids
- Sterol lipids
- Prenol lipids
- Saccharolipids
- Polyketides
Each lipid heads its own subclassification hierarchy, according to the classification system proposed by LIPID Metabolites and Pathways Strategy (LIPID MAPS, La Jolla, California, USA; www.lipidmaps.org).
Several websites provide useful overviews of lipid structure and function, as well as analytical procedures for lipid analysis: Lipid Library, lipidlibrary.co.uk; Lipid Bank, lipidbank.jp; and Cyberlipids, www.cyberlipid.org.
Parallel to the development of new technologies is how our understanding of the biological role of lipids has changed with time. Lipids were known to serve as the structural backbone of cell membranes and as storage for metabolic energy. Recently, the biomedical community learned that lipids play pivotal roles in regulating a wide variety of cellular processes in all organisms. Within each cell exist thousands of types of lipids whose composition, or lipid profile, changes in response to chemical signals from the cell's environment. Studying lipid profiles can provide insight to certain health and disease processes.
Comprehensive analysis of a wide array of lipids in biological samples is a challenge primarily for analytical chemistry. These complex mixtures of lipids have a large variety of chemical structures and a large, dynamic range of concentrations. Consequently, interest in adapting novel technologies for lipid analysis continues undiminished.
100 Years of Lipid Analysis
Our knowledge of lipid biology has been hampered by analytical limitations. At the beginning of the 20th century, lipid analysis was based largely on gravimetric measurements. The use of radioactive and colourimetric assays allowed the initial characterization of lipid metabolism and the understanding that one lipid species could be interconverted into another one. In the 1950s, lipids were used as molecule tools to probe the basic mechanisms of ion fragmentation following electron ionization (EI) (4,5). As such, they provided a glimpse of the potential of MS in biochemical applications.
By the 1960s, the application of thinlayer chromatography (TLC) made the separation of the complex mixture of lipids present in biological samples possible.
Identifying the specific fatty-acyl substituents in glycerophospholipids involved separating lipid classes by TLC, scraping the TLC spots, extracting and hydrolyzing the fattyacyl group, and derivitizing for gas chromatography–mass spectrometry (GC–MS). Although still used today, this approach often requires the extraction of a fairly large amount of lipids. Moreover, the derivatization process renders analyses highly labour-intensive and insufficiently sensitive to target lipids present at low concentrations in biological tissues.
In the 1980s, with the development of fast-atom-bombardment mass spectrometry (FAB-MS), we could directly analyse nonvolatile lipids, such as glycerophospholipids, as intact molecules, preserving the information inherent in their chemical structure. We could also determine the position of the fatty acyl substituents on the glycerol backbone of glycerophospholipids and even define the position of double bonds present within the fatty-acyl chain through remotesite fragmentation mechanisms. Nevertheless, fragmentation of the lipid analyte during ionization and the high chemical backgrounds observed in early studies with FAB were disadvantages, in particular for the analysis of complex lipid mixtures, making the interpretation of the lipid spectra a tedious task.
The development in the late 1980s of electrospray ionization (ESI) (see sidebar "Solvent-Based Ionization Techniques"), for which John Fenn shared the 2002 Nobel Prize in Chemistry, revolutionized the study of lipid biology. ESI enabled the introduction of nonvolatile lipid species from the liquid phase into the gas phase directly, without the need for prior derivatization. Furthermore, ESI greatly simplifies lipid analysis. It is a "soft" ionization technique, which, compared with FAB-MS, results in decreased molecular ion decomposition, better reproducibility and lower detection limits. Because this soft ionization technique does not fragment lipids, we can correlate comprehensive detection of an entire range of lipids within a complex mixture to experimental conditions or disease state.

Solvent-Based Ionization Techniques
A particular advantage of ESI lies in its ability for coupling to an LC system, making the analysis highly sensitive and quantitative. Also, because of its enclosed design, LC minimizes the exposure of lipids to atmospheric oxygen, reducing sample oxidation relative to that occurring during TLC separations.
The parallel development of MS techniques lends itself to lipid analysis. Those techniques are tandem MS (MS-MS) and the highmass-resolution instruments, which include triple quadrupole, ion trap, time-of-flight (TOF), Fourier-transform ion cyclotron resonance (FT-ICR) and orbital trap (Orbitrap, Thermo Fisher Scientific, Waltham, Massachusetts, USA). MS proved itself a sensitive tool for identifying, characterizing and quantifying lipid species. Therefore, it replaced some of the immunological, radioactive, fluorometric and colourimetric techniques used in traditional laboratory analysis.
Today, mass spectrometry, a term whose meaning encompasses sample preparation, chromatography, ionization and mass analyzers, provides the tools to simultaneously measure the biological levels of thousands of lipids.
Modern Lipid Analysis: Lipidomics
In the 2000s, the development of new approaches for MS-based lipid analysis led to the emerging field of lipidomics (6–8) (Figures 2 and 3). Lipidomics is more than just the decades-old lipids analysis performed with new technological tools. It is a powerful new way to observe and describe lipid biology. We define lipidomics as the discipline that studies the largescale changes in lipids accompanying perturbations of biological systems or specific phenotypes. Because lipids constitute a subset of metabolites (including water-soluble components such as amino acids, carbohydrates and nucleotides), lipidomics is considered part of metabolomics (Figure 2). Nevertheless, the distinct solubility properties of most lipids (waterinsoluble) dictate their separate analysis in metabolomic experiments. Notably, the lipid composition of a biological sample reflects both genetic and environmental components (Figure 2), which differentiates lipidomics from the other "–omics" disciplines (genomics, transcriptomics and proteomics).
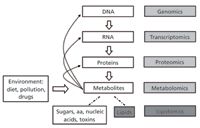
Figure 2: Lipidomics, the systems-level scale analysis of lipids and their interacting partners, can be viewed as a subdiscipline of "metabolomics," under the umbrella of systems biology. Abbreviations: aa = amino acids.
A typical lipidomic experiment often begins with extracting lipids from tissues or cells (Figure 4). The complex lipid mixture is then analysed with an MS system. A lipid profile results, yielding information about the lipid composition of the starting material and the abundance of individual lipids it contains. Typically, such experimental readouts are referenced to a control condition, to deduce distinct lipid species whose levels change upon perturbation of the biological system under investigation, which may include diseases, nutritional or pharmacological treatments, environmental stresses and genetic manipulations.
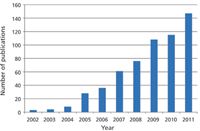
Figure 3: Exponential growth in the number of publications containing the term "lipidomics".
Generating comprehensive profiles of the lipids present in biological samples presents a major challenge for lipidomics. The samples vary in concentration (from attomolar to nanomolar) and chemical complexity (thousands of components). Complex analytical strategies study lipid phenotypes as well as perform comparative analysis of lipidomes, which is the entire set of lipids present in an organism. Currently, we are pursuing complementary strategies for lipidomic investigation: targeted lipidomics and untargeted lipidomics.
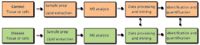
Figure 4: Flow chart of a lipidomic analysis.
Targeted lipidomics is a hypothesisdriven approach that focuses on analysing a selected group of lipids. These lipids can be related to a specific class or metabolic pathway, a particular set of biomarkers or substrate and product of lipid enzymatic reactions (9–13). The specific lipids that will undergo analysis are selected according to the questions asked, and ad hoc analytical methods are developed for the lipids' absolute quantification.
Technological advancements, however, help increase the number of lipids that we can simultaneously quantify in a single analysis. This increase enables us to measure lipids from different biochemical pathways, our objective being to quantify the entire lipidome in a more classical "-omic" style. Today, using selected retention times with ultrahighpressure liquid chromatography (UHPLC) together with multiple-reaction monitoring (MRM) transitions and triplequadrupole instruments, we can monitor several hundred lipid species in a single LC–MS analysis (9–13).
Untargeted lipidomics, the second approach, is a typical "-omic" approach. As such, it is hypothesisgenerating and exploratory in nature; its purpose is not to identify each observed lipid but rather to perform an unbiased screening of lipids, obtaining profiles or "fingerprints" of as many lipids as possible in biological samples. By comparing lipid profiles, we can thus determine patterns of variations between different groups: healthy versus diseased, control versus treated and wild-type versus genetically modified (14–17). Dealing with variations in thousands of molecular species, untargeted lipidomic strategy applies statistical tools such as computational sorting, multivariate analysis and pattern-recognition analysis to group the observed changes in lipid species.
For untargeted lipidomics, mass spectrometers that offer high resolution and high accuracy are generally used, including TOF, FT-ICR and orbital trap systems (14–17). Interestingly, because untargeted lipidomics lead to the detection of unexpected or unknown lipids in biological samples, elucidating their chemical structures often relies on MS-MS or multiple fragmentation (MSn ) capabilities. In this context, datadependent acquisition (DDA) and data-independent acquisition modes (see sidebar "Common Fragmentation Experiments in Lipidomics as Performed on Ion-Trap and TandemHybrid Instruments") obviate the need to analyse the sample in MS mode and then rerun the same sample in MS-MS mode, thus acquiring the fragmentation data from each of the selected precursors (14–16). For that reason, tandem hybrid instruments based on TOF, FT-ICR and orbital trap technologies could provide simultaneously qualitative information on the number and structures of the components present in a mixture, as well as quantitative information. The simultaneous generation of highresolution, full-scan and fragmentation spectra could serve as a repository of biological information that can be reused repeatedly with new knowledge.

Common Fragmentation Experiments in Lipidomics as Performed on Ion-Trap and Tandem-Hybrid Instruments
Lipidomics in Research Labs
Currently, lipidomics is a routine strategy in research laboratories and a powerful new tool in systems biology. Together with genomics, transcriptomics and proteomics, lipidomics is being used in scientific areas such as disease diagnostics, drug discovery, drug development, personalized medicine, biotechnology and metabolic engineering.
The role of lipidomics in diagnostics helps researchers compare the lipidomes of healthy individuals with those of diseased subjects. Indeed, lipid composition can provide a "snapshot" of the biological state of an organism. It is, consequently, an index, or biomarker, of a healthy or diseased state. In addition, lipidomics can be used to monitor the outcome of treatment strategies, such as pharmacological or dietary interventions, by observing whether the lipidome of treated and diseased patients shifts in the cluster of healthy subjects.
Regarding drug discovery, the unbiased screening of lipid changes accompanying a disease state could lead to the discovery of novel, unexpected biological pathways involved in disease etiology, suggesting new targets for therapeutic intervention.
A detailed knowledge of the pharmacological properties of drugs is essential for the development of new pharmaceuticals. Such knowledge is often required for assessing the benefits of new drugs on the market. An increased focus on safety and toxicology and differentiating new treatments from existing ones has created an environment where lipidomics could be used as an approach for obtaining a detailed picture of the effect of a drug on the body, enabling an assessment of the complex biological response to new chemical entities.
The concept of personalized medicine, a model for future health care delivery, includes both nutritional and pharmacological treatments personalized to metabolic phenotypes. It will rely strongly on lipidomics.
Many scientific investigations are conducted using plant, yeast, bacteria or animal cells as research models. In these simple biological systems, lipid profiles can be regarded as the ultimate response to genetic or environmental changes (Figure 2). Hence, lipidomics can be used to determine the function of genes and proteins after genetic manipulation: gene overexpression, gene knockdown and transgene. It can also provide the basis for investigating the effects of genetic diversity (genotypes, epigenetic regulation, mutations and polymorphism) and environmental stresses such as chemicals, climate, radiation, toxins and nutrients on lipid metabolism.
Conclusions
In the past decade, a series of technological advancements in the field of MS-based lipid analysis gave rise to lipidomics, a subspecialty of metabolomics. Lipidomics is already revolutionizing lipid research. We can now amass certain types of experimental data about lipid composition 100 times or, in some cases, 1000 times faster than we could 20 years ago. The resultant volume of information offers an unprecedented opportunity to describe biological systems and thus lend itself to unexpected scientific discoveries.
Novel MS technology is being adopted to analyse the wide array of lipids present in biological samples. In Part 2 of this column, I will describe the use of such technology for lipidomics in more detail.
Acknowledgments
The author would like to thank Dave Sarro for providing helpful advice and editorial support.
Giuseppe Astarita is principal scientist in lipidomics and metabolomics at Waters (Milford, Massachusetts, USA) and serves as an adjunct professor at Georgetown University (Washington, D.C., USA). An expert in lipid MS and lipidomics for translational medicine and biomarker discovery, his work appears in several peer-reviewed scientific publications and book chapters. The International Society for the Study of Fatty Acids and Lipids has bestowed upon Dr Astarita the Top New Investigator Award in Biochemistry of Lipids. Direct correspondence to: giuseppe_astarita@waters.com.
"MS — The Practical Art" Editor Kate Yu joined Waters in Milford, Massachusetts, USA, in 1998. She has a wealth of experience in applying LC–MS technologies into various application fields such as metabolite identification, metabolomics, quantitative bioanalysis, natural products and environmental applications. Direct correspondence about this column should go to "MS: The Practical Art", LCGC Europe, 4A Bridgegate Pavilion, Chester Business Park, Wrexham Road, Chester, CH4 9QH, UK or e-mail the editor, Alasdair Matheson, at amatheson@advanstar.com
References
(1) O. Quehenberger and E.A. Dennis, N. Engl. J. Med., 365(19), 1812–1823 (2011).
(2) M.R. Wenk, Nat. Rev. Drug Discov., 4(7), 594–610 (2005).
(3) E. Fahy et al., Lipid Res., 46(5), 839–861 (2005).
(4) R. Ryhage and E. Stenhagen, J. Lipid Res., 1, 361–390 (1960).
(5) G.E. VanLear and F.W. McLafferty, Annu. Rev. Biochem., 38, 289–322 (1969).
(6) M. Bou Khalil et al., Mass Spectrom. Rev., 29(6), 877–929 (2010).
(7) R.C. Murphy and S.J. Gaskell, J. Biol. Chem., 286(29), 25427–25433 (2011).
(8) G. Astarita et al., Methods Mol. Biol., 579, 201–219 (2009).
(9) G. Astarita and D. Piomelli, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci., 877(26), 2755–2767 (2009).
(10) C.A. Haynes et al., J. Chromatogr. B Analyt. Technol. Biomed. Life Sci., 877(26), 2696–2708 (2009).
(11) C.N. Serhan, Prostaglandins Other Lipid Mediat., 77(1–4), 4–14 (2004).
(12) T.D. Niemoller and N.G. Bazan, Prostaglandins Other Lipid Mediat., 91(3–4),85–89 (2009).
(13) M. Balazy, Prostaglandins Other Lipid Mediat., 73(3-4), 173–180 (2004).
(14) X. Han and R.W. Gross, Mass Spectrom. Rev., 24(3), 367–412 (2005).
(15) M. Ståhlman et al., J. Chromatogr. B Analyt. Technol. Biomed. Life Sci., 877(26), 2664–2672 (2009).
(16) J.M. Castro-Perez et al., J. Proteome Res., 9, 2377–2389 (2010).
(17) K. Schuhmann et al., Anal. Chem., 83(14), 5480–5487 (2011).


.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)










