Characterization of PLGA Using SEC–MALS-VIS
The Application Notebook
Poly(lactic-co-glycolic acid) (PLGA) is a copolymer based on glycolic acid and lactic acid. The two monomer units are linked together by ester linkages and form linear polyester chains
Poly(lactic-co-glycolic acid) (PLGA) is a copolymer based on glycolic acid and lactic acid. The two monomer units are linked together by ester linkages and form linear polyester chains. The obtained product is biodegradable and biocompatible, and it is approved by the Food and Drug Administration (FDA) for production of various therapeutic devices as well as for drug delivery applications. The properties of PLGA can be tuned by the ratio of the two monomers and by its molar mass distribution.
The characterization of PLGA by means of conventional size-exclusion chromatography (SEC) is problematic because of the lack of suitable calibration standards. In addition, the linear polyester structure can be modified by the addition of small amounts of polyfunctional monomer to obtain branched chains of differing degrees of branching. The degree of branching becomes an additional parameter that can be used to adjust PLGA properties-all of which renders conventional column calibration an inadequate analytical technique.
In this application note, two commercially available samples were analyzed by SEC coupled to a multi-angle light scattering (MALS) detector (HELEOS), a refractive index detector (Optilab rEX), and a viscosity (VIS) detector (ViscoStar). The ViscoStar was used in order to discover additional information about the molecular structure of the analyzed polymers. In addition to molar mass distributions, the SEC–MALS-VIS system yields the relationship between intrinsic viscosity and molar mass (Mark-Houwink plot) that can provide deep insight into the molecular structure of the polymers being analyzed.
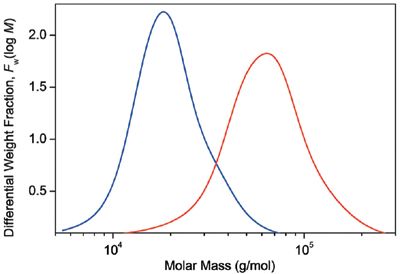
Figure 1: Differential molar mass distribution curves of two PLGA samples.
In Figure 1, the molar mass distributions are given as differential distribution plots. As seen from the plots, the two samples span markedly different molar mass ranges. The Mark-Houwink plots of the two samples are shown in Figure 2 together with the plot of linear polystyrene that is shown simply for the sake of comparison. The slope of the Mark-Houwink plot of the linear polystyrene is 0.71, a typical value for linear random coils in thermodynamically good solvents. The slope of the red sample roughly corresponds to a linear structure as well. However, there is a slight indication of deviation from linearity at the region of high molar masses that may indicate the presence of branched molecules. The Mark-Houwink plot of the blue sample is curved. Curvature of the Mark-Houwink plot generally reveals branching. In addition, the slope of the higher molar mass portion of the Mark-Houwink plot of 0.48 suggests significant branching.
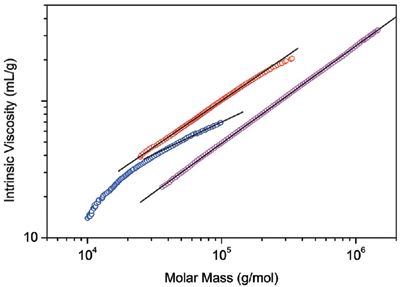
Figure 2: Mark-Houwink plots of two samples of PLGA (red and blue) and linear polystyrene (magenta). The lines are linear extrapolations of the data.
SEC-MALS-VIS is an excellent method for the characterization of PLGA polyesters as it has the ability to determine not only the molar mass distribution, but to reveal subtle differences in PLGAs molecular structure.

Wyatt Technology Corporation
6300 Hollister Avenue, Santa Barbara, CA 93117
tel. (805) 681-9009, fax (805) 681-0123
Website: www.wyatt.com

SEC-MALS of Antibody Therapeutics—A Robust Method for In-Depth Sample Characterization
June 1st 2022Monoclonal antibodies (mAbs) are effective therapeutics for cancers, auto-immune diseases, viral infections, and other diseases. Recent developments in antibody therapeutics aim to add more specific binding regions (bi- and multi-specificity) to increase their effectiveness and/or to downsize the molecule to the specific binding regions (for example, scFv or Fab fragment) to achieve better penetration of the tissue. As the molecule gets more complex, the possible high and low molecular weight (H/LMW) impurities become more complex, too. In order to accurately analyze the various species, more advanced detection than ultraviolet (UV) is required to characterize a mAb sample.















