Waters - An Anion-Exchange Chromatography Method for Monitoring Empty Capsid Content in Adeno-Associated Virus Serotype AAV8
The single-stranded DNA (ssDNA) content of AAV capsids in AAV-based gene therapy preparations impacts the efficacy of this treatment modality. We demonstrate that AAV8 capsids without and with full length ssDNA can be separated and their relative abundances determined on a Waters Protein-Pak Hi Res Q strong anion-exchange column.
During the recombinant adeno-associated virus (AAV) biomanufacturing process, capsids that do not contain ssDNA (empty capsids) are also produced. Empty capsids are unable to deliver ssDNA to the targeted cells and as a result are important to monitor during process development and product manufacturing. An anion-exchange chromatography (AEX) separation driven by differences in surface charge can monitor changes in empty capsid levels (1,2). AEX using fluorescent detection consumes small sample amounts and can provide reliable and reproducible results.
Experimental Conditions
Samples: AAV8-CMV-GFP (full capsid) and AAV8-null (empty capsid) 2 × 1012 capsids/mL; other AAV-CMV-GFP serotypes: ranging from 1.3 × 1013 to 6.7 × 1013 GC/mL = genome copies (physical titer)/mL
LC system: ACQUITYTM UPLCTM H-Class Bio
Sample temp.: 10 ̊C
Column temp.: 30 ̊C
Injection volume: 0.2–6 μL
Column: Waters Protein-Pak Hi Res Q, 4.6 × 100 mm
Fluorescence detector: Excitation: 280 nm; Emission: 350 nm
Gradient: 70 mM bis-tris propane, pH 9.0, 100–300 mM tetramethylammonium chloride in 20 min at 0.4 mL/min
Results and Discussion
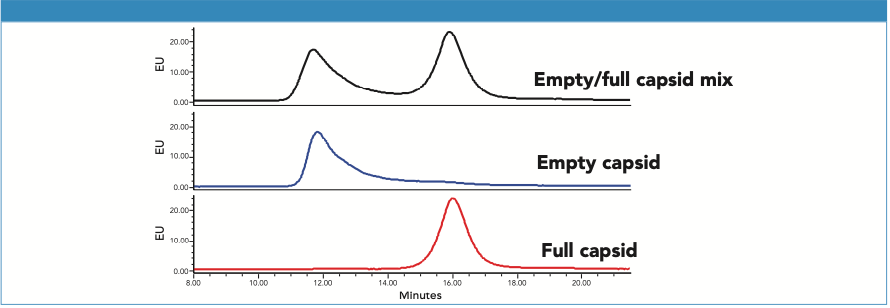
The AEX separation of empty and full AAV8 capsid was optimized for several parameters including pH and salt types (3). In addition, intrinsic protein fluorescence was monitored to provide greater sensitivity and selectivity, and less baseline drift (2). The separation achieved is shown in Figure 1. We observe that the ssDNA “empty” capsids are partially coelute with the ssDNA “full” capsids while showing no significant change in the UV absorbance ratio (280 nm/260 nm, data not shown), indicating the unlikelihood of monitoring AAV8 with partial ssDNA by AEX.
Figure 1: AAV8 ssDNA (CMV-GFP) empty and full capsids are separated on a Protein-Pak Hi Res Q Column using an optimized AEX method (see experimental conditions for method details).
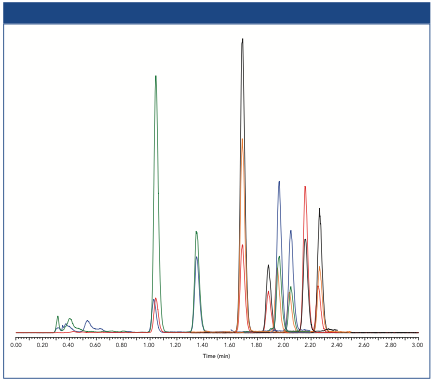
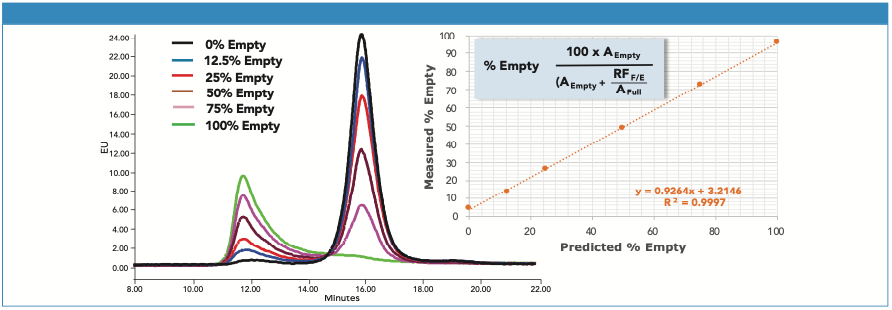
A series of chromatograms for AAV8 samples with differing percentages of empty capsids is presented in Figure 2. The mixtures were generated from >99% pure “empty” and “full” samples. The sample concentrations were normalized based on FLR peak areas from a size-exclusion chromatography (SEC) separation with fluorescence detection and assumed a “full” to “empty” fluorescence response factor (RFF/E) of 1.3 (2). For AEX, however, 1.9 was calculated for RFF/E (Area100% Full/Area100% Empty). The discrepancy in RFF/E values between SEC and AEX may be the result of changes in quantum efficiency under different mobile-phase conditions. RFF/E is then divided into the full capsid peak area to calculate the percentage of empty capsid in a sample (Figure 2). The accuracy of this approach depends on the veracity of RFF/E, which requires reasonably pure “full” and “empty” samples, and accurate capsid concentrations. While RFF/E was not rigorously defined in this work, the estimate of RFF/E used here effectively demonstrates the approach to interpretation of the AEX data. Determining RFF/E is not essential, if only relative comparisons of empty capsid levels between samples are required.
Figure 2: Shown on the left is an overlay of AEX chromatograms for AAV8 samples with differing levels of empty capsid. On the right is the correlation between predicted and measured empty capsid content. Calculation of the percentage of empty capsid was based on the equation shown, which corrects for the difference in fluorescence response between empty and full capsids. AEmpty and AFull are the AEX peak areas of the empty and full capsids, and RFF/E is the fluorescence response factor.
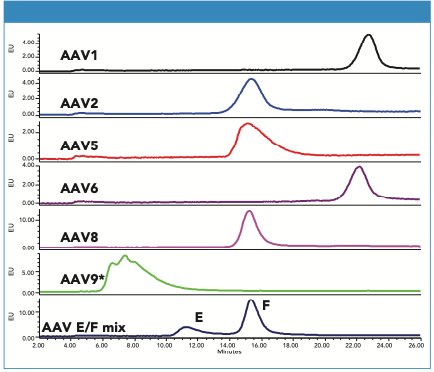
Samples of several AAV-CMV-GFP serotypes were also evaluated (Figure 3). All tested serotypes were retained on the column, but their retention varied significantly.
Figure 3: Chromatograms of AAV1, AAV2, AAV5, AAV6, AAV8, and AAV9 serotypes (full capsids, CMV-GFP ssDNA). For comparison, the bottom chromatogram shows the optimized separation of AAV8 empty and full capsid. Separation conditions are as described in the text for all serotypes except for AAV9 (*) where the salt gradient was 0–200 mM KCl.
Conclusions
AEX will not likely provide a measurement of AAV8 capsid carrying partial ssDNA, therefore complementary methods such as analytical ultracentrifugation (AUC) would be needed for that analysis. However, due to the technical challenges and costs of these methods, AEX may still provide utility as an initial screening tool in support of manufacturing process development and as a more precise assessment of changes in empty capsid content in product quality testing.
Due to differences in capsid protein surfaces, method optimization may be needed when separating empty and full capsids of other AAV serotypes, and the AEX method described here can serve as a useful starting point.
References
(1) X. Fu, et al, Hum. Gene. Ther. Methods 30(4), 144 (2019).
(2) C. Wang, et al, Mol. Ther. Methods Clin. 19, 257 (2019).
(3) H. Yang, et al, Waters Application Note 720006825EN (2020).
Waters, The Science of What’s Possible, Protein-Pak, ACQUITY, and UPLC are trademarks of Waters Corporation. All other trademarks are the property of their respective owners.

Waters Corporation
34 Maple Street, Milford, MA 01757
tel. 508 478 2000
Website: www.waters.com

SEC-MALS of Antibody Therapeutics—A Robust Method for In-Depth Sample Characterization
June 1st 2022Monoclonal antibodies (mAbs) are effective therapeutics for cancers, auto-immune diseases, viral infections, and other diseases. Recent developments in antibody therapeutics aim to add more specific binding regions (bi- and multi-specificity) to increase their effectiveness and/or to downsize the molecule to the specific binding regions (for example, scFv or Fab fragment) to achieve better penetration of the tissue. As the molecule gets more complex, the possible high and low molecular weight (H/LMW) impurities become more complex, too. In order to accurately analyze the various species, more advanced detection than ultraviolet (UV) is required to characterize a mAb sample.