Hilicon AB - Tailored Analysis of Phospholipid Classes Using iHILIC-Fusion(+) as First Dimension for Online Two-Dimensional HILIC–Reversed-Phase LC–MS
The Application Notebook
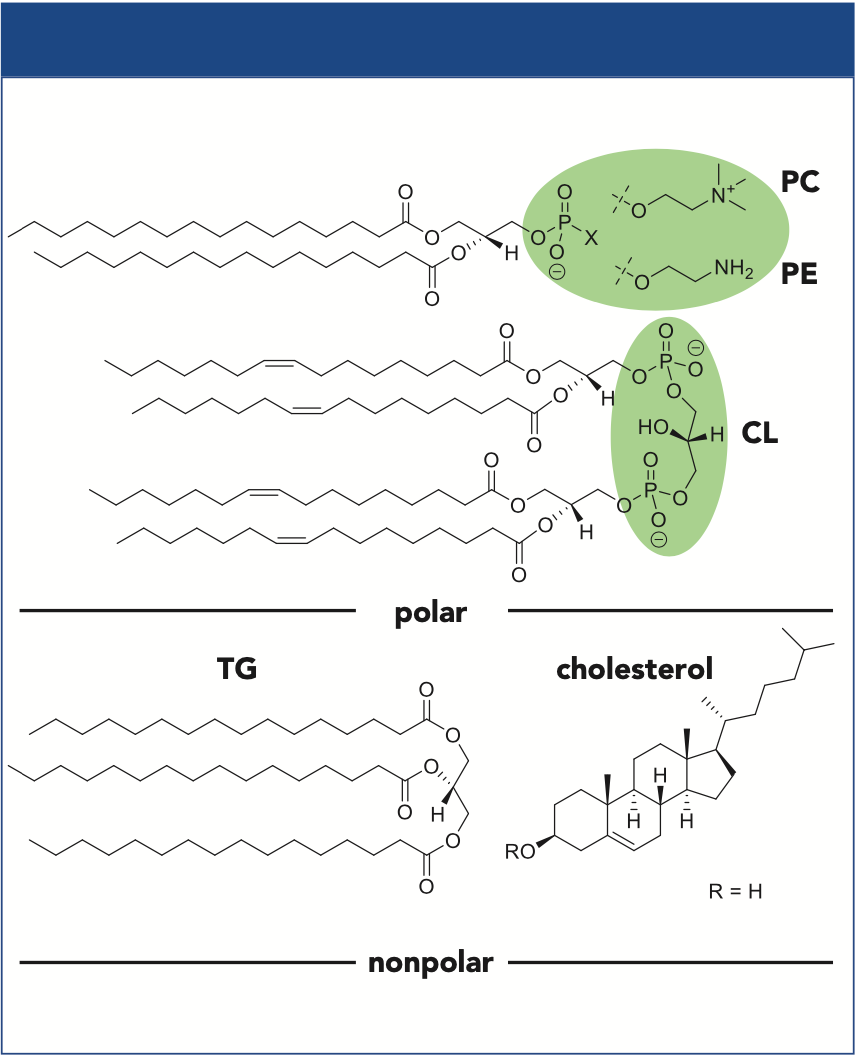
Phospholipids (PLs) are a large lipid subgroup that is involved in important cell functions in all organisms. They are the main components of biological membranes and are essential for many biologic processes within and between cells, for example, signal transduction, cell growth, and various transport processes. The basic building blocks for PLs are fatty acids linked to a glycerol backbone and polar head groups (Figure 1). In addition to the main membrane PLs, such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE), other PL classes are of great importance as well. Some PLs have significantly lower concentrations. For example, in eukaryotic organisms, cardiolipin (CL) is exclusively located in the inner mitochondrial membrane (1). Thus, the concentration of CL in total lipid extracts is very low in comparison to the major membrane lipids, such as PLs and cholesterol, but also triacylglycerols (TGs) (2,3). Furthermore, ion suppression effects complicate the mass spectrometric (MS) analysis, and a tailored analysis of low abundant PL classes is often required.
Figure 1: Selected structures of nonpolar lipids and polar PLs. HILIC separation of PLs takes place according to their polar head groups (shown in green).
In our previous study, we demonstrated the advantage of performing sample preparation by solid-phase extraction (SPE) in hydrophilic interaction liquid chromatography (HILIC) mode (4). The nonpolar fraction of lipids was first separated from polar PLs using an offline HILIC SPE method, and thereafter separation by reversed-phase liquid chromatography (LC) was performed. Offline methods often require solvent evaporation and reconstituting analytes in a suitable solvent for the second dimension, which is a time-consuming approach and risks losing labile PLs. In this application, a novel online heart-cut two-dimensional (2D) HILIC–reversed-phase LC–MS method is presented for the separation of PL classes and nonpolar lipids, where HILIC was utilized in the first dimension (1D) and reversed-phase LC in second dimension (2D).
Experimental
Lipid Standards: Triacylglycerol (TG 48:0), cholesterol (Chol), phosphatidylcholine (PC 32:0), phosphatidylethanolamine (PE 32:0), cardiolipin (CL 64:4), and yeast total lipid extract (Saccharomyces cerevisiae).
LC–MS/MS Setup: Thermo Scientific Ultimate 3000 system with dual gradient pump hyphenated to a Q ExactiveTM Plus Hybrid Quadrupole-OrbitrapTM mass spectrometer. Ionization was performed utilizing electrospray ionization in negative ionization mode as described by Helmer et al. (3).
1D HILIC Separation:
Column: 20 × 2.1 mm, 5-μm, 100 Å, iHILIC®-Fusion(+) (P/N 100.022.0510, HILICON)
Eluents: A) ammonium formate solution (35 mM, pH 3.5) and 95:5 (v/v) acetonitrile; B) acetonitrile
Gradient Elution: 0–0.2 min, 97% B; 0.2–0.5 min, from 97% to 93% B; 0.5–2.75 min, 93% B; 2.75–7.5 min, from 93% to 60% B; 7.5–11 min, 60% B; 11–11.5 min, from 60% to 97% B; re-equilibration is parallel to reversed-phase LC separation at 97% B.
Flow Rate: 0.3 mL/min; 0.05 mL/min (during parallel reversed-phase LC separation)
Column Temperature: 40 °C
Injection Volume: 2–20 μL
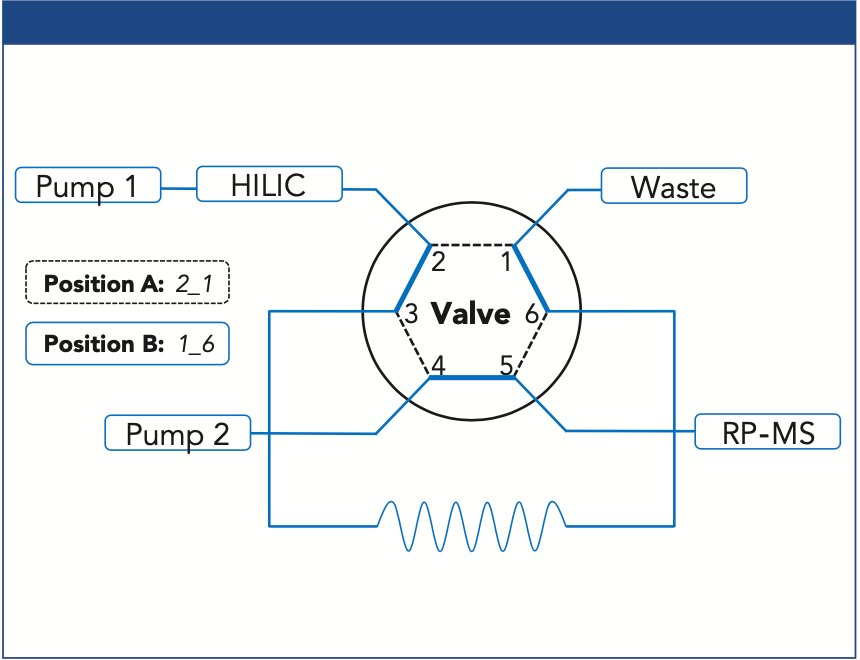
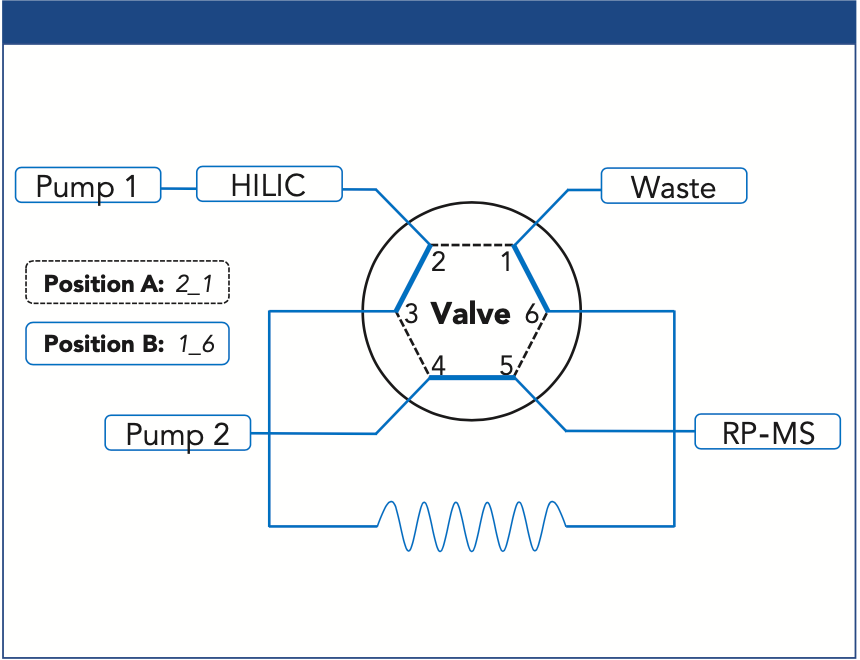
Heart-Cut Setup for PL Transfer: An online heart-cut 2D HILIC–reversed-phase LC–MS setup was developed for PL transfer from 1D (HILIC) to 2D (reversed-phase LC). As described earlier, a six-port valve equipped with a sample loop (300 μL, PEEK) was utilized as the transfer (Figure 2) (3). At first, the HILIC eluate was directed to the waste. By switching the valve from A (2_1) to B (1_6) position, the selected PL fraction was transferred into the loop for the separation in the second dimension. Since we focused on CL in this study, the transfer time window was set at its retention time of 6.35–6.95 min.
Figure 2: Valve setup for the transfer of PL classes from 1D HILIC to 2D reversed-phase (RP) LC via heart-cut approach.
Results and Conclusion
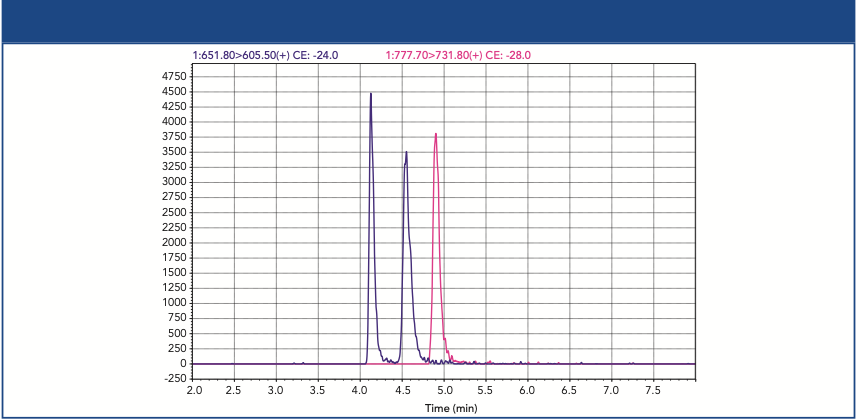
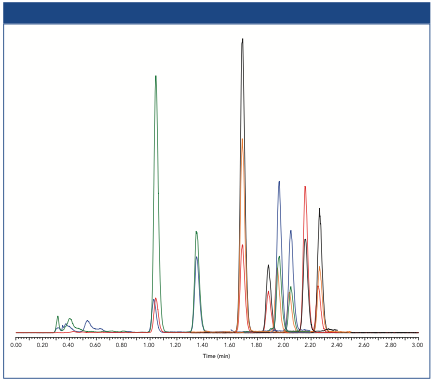
A tailored analysis of CL species in yeast (S. cerevisiae) was achieved utilizing two-dimensional HILIC–reversed-phase LC–MS. In the first dimension, the separation of PL classes was carried out using a HILIC method. In addition to that, nonpolar lipids did not show retention and were eluted at the void volume. As shown in Figure 3(a), the lipid classes (TG, PE, CL, and PC) were well separated according to their different head groups with this new method. The application of this method to a yeast total lipid extract (Figure 3[b]) shows the intended coelution of the CL species in yeast, where one typical CL species is CL 66:4 with 66 carbon atoms and four double bonds in the four linked fatty acids.
Figure 3: (a) 1D HILIC separation of representative lipid standards; (b) coelution of dominant CL species in yeast (S. cerevisiae) within the transfer window; (c) comparison of reversed-phase LC TICs in 1D (red) and 2D (black).
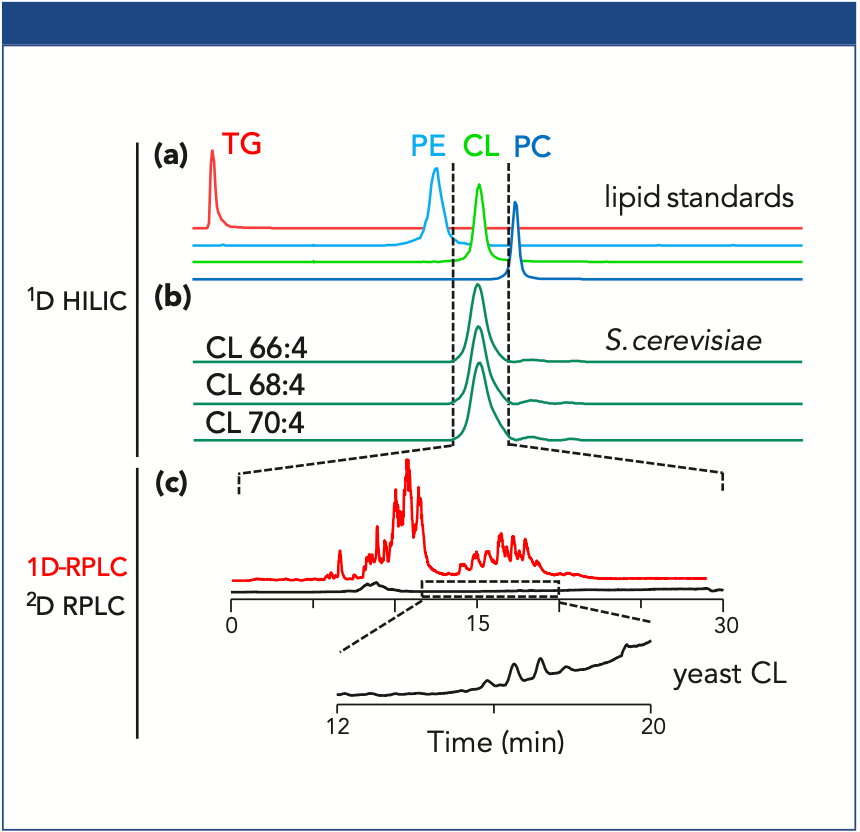
In the developed 2D HILIC–reversed-phase LC separation method, a transfer window with all CL species from 1D HILIC was first determined for the transfer via a heart-cut setup to the 2D reversed-phase LC system. Compared to the conventional one-dimensional (1D) reversed-phase LC (red), 2D reversed-phase LC (black) shows a significant decrease of background signals (mainly TGs), and low abundant CL species can even be observed in the total ion current (TIC) chromatogram (Figure 3[c]). Furthermore, by the reduction of the background as observed in 1D reversed-phase LC, higher CL signal intensities have been observed (3).
The selective HILIC pre-separation of lipid classes in the first dimension significantly reduced the matrix effects in the second dimension separation of low abundant PL classes. Interestingly, low abundant CL species can be sensitively detected and observed, even in the TIC. In summary, the automated online 2D method via heart-cut transfer offers a fast and gentle sample preparation and clean-up procedure that is very useful for analyzing low abundant PL classes as well as labile PLs, such as PL oxidation products (3).
References
(1) J.M. Berg et al., Stryer Biochemistry (Springer), (2018).
(2) M. Lange et al., Chromatographia 82, 77–100 (2018).
(3) P.O. Helmer et al., J. Chromatogr. A. 460918, in press https://doi.org/10.1016/j.chroma.2020.460918
(4) P.O. Helmer et al., Rapid Commun. Mass Spectrom. 34, e8566 (2020). https://doi.org/10.1002/rcm.8566

Hilicon AB
Tvistevägen 48, SE-90736 Umeå, Sweden
Tel.: +46 (90) 193469
E-mail: info@hilicon.com
Website: www.hilicon.com

MALDI Guided SpatialOMx® Uncovers Proteomic Profiles in Tumor Subpopulations of Breast Cancer
September 1st 2020The timsTOF fleX system bridges a current gap by providing MALDI Imaging and in-depth proteomics analysis in just one instrument. The instrument offers all benefits of a timsTOF Pro for time-efficient and sensitive proteomics, combined with a high-resolution MALDI source and stage. Using PASEF technology, it is possible to retrieve high protein ID rates with small sample amounts. Here we present the new SpatialOMx® workflow to identify distinct proteomic profiles for different tumor subpopulations in breast cancer as an example for this powerful approach.