Trends in Bioanalysis Using LC–MS–MS
Bioanalysis of biologics presents a number of technical challenges. Ligand binding assays (LBA) are the gold standard bioanalytical technique for quantification of biologics in complex matrices such as serum and plasma but selectivity issues and the need for specific capture reagents limit their applicability in the drug discovery and development phase. Liquid chromatography coupled with tandem mass spectrometry (LC-MS-MS) is widely used for highly selective and sensitive bioanalysis of small molecules. However, large molecule bioanalysis presents challenges including the need for extensive and complex sample preparation for LC-MS-MS. This article explores the limitations of LC-MS-MS for bioanalysis of biologics and some of the latest trends for overcoming these in bioanalysis laboratories.
Suma Ramagiri, Sciex, Toronto, Canada.
Photo Credit: merrymoonmary/Getty Images

Bioanalysis of biologics presents a number of technical challenges. Ligand binding assays (LBA) are the gold standard bioanalytical technique for quantification of biologics in complex matrices such as serum and plasma but selectivity issues and the need for specific capture reagents limit their applicability in the drug discovery and development phase. Liquid chromatography coupled with tandem mass spectrometry (LC-MS-MS) is widely used for highly selective and sensitive bioanalysis of small molecules. However, large molecule bioanalysis presents challenges including the need for extensive and complex sample preparation for LC-MS-MS. This article explores the limitations of LC-MS-MS for bioanalysis of biologics and some of the latest trends for overcoming these in bioanalysis laboratories.
The biologics market has grown rapidly in recent years, with several new therapeutics being approved annually.1 Traditional approaches for quantification of biologic drugs typically involve either ligand-binding assays (LBAs) such as enzyme-linked immunosorbent assays (ELISA), or UV detection of individual peptides using high performance liquid chromatography (HPLC) separation. LBAs rely on immunoaffinity detection of a unique epitope on the protein or peptide of interest. The high specificity of the antibody-based interactions can track an analyte at high sensitivity, although the dynamic range is narrowed to just one or two orders of magnitude.2 LBAs can detect both physiologically active and “free” forms of circulating large molecule drugs in samples. They are popular for their ease of implementation, but are typically expensive and time-consuming to develop,3 and are subject to limited selectivity and antibody cross-reactivity. This results in lack of specificity from interference, and high background levels that are not suitable for meeting the biopharmaceutical industry’s requirements to detect diverse proteins and peptides with ever-increasing sensitivity and reproducibility.
Since the 1980s, liquid chromatography coupled with tandem mass spectrometry (LC-MS-MS) has been used extensively in pharmaceutical laboratories for small molecule bioanalysis, and is favoured for its high throughput, sensitivity, accuracy, and selectivity. LC-MS-MS has similar advantages for biologics; importantly, it is not subject to antibody cross-reactivity because LC-MS-MS involves direct evaluation of the chemical properties of the analyte.2 LC-MS-MS also offers excellent selectivity, being able to distinguish and quantify highly homologous isoforms - even at low levels - with accuracy and precision over a wide linear dynamic range.4
However, performing LC-MS-MS-based bioanalysis for large molecule drugs raises a number of new challenges that can be especially difficult for laboratories switching from small to large molecule workflows. In particular, sample preparation and extraction steps for quantification of large molecules can be complicated and very laborious to optimize because bioanalysis samples are highly complex biological matrices that contain numerous background peptides and proteins. Furthermore, the analysis of such samples is more challenging, because the background peptides and proteins compete with the biotherapeutic molecule of interest, creating interference problems and impacting on accuracy. LC-MS-MS approaches are also unable to capture free drugs that may be circulating in serum, thereby missing important data during quantification.
As a result, bioanalysis laboratories face several common issues when studying biologics, including insufficient selectivity amid background interference; insufficient sensitivity at low levels of detection (picogram quantities); and sub-standard data quality and reproducibility, associated with the challenges of quantifying peptides and proteins. These issues have driven several new technology and method developments, including improvements in sample preparation, detector technologies and sensitivity, and selection methods.
Improving Sample Preparation and Selectivity
Quantification of large molecules requires sample extraction, which typically involves one of several different immunocapture approaches. These can be drug target-specific, using an antibody developed for the specific drug of interest; target specific, using receptor-specific antibodies to the drug molecule’s complementarity determining region (CDR); or semi-targeted capture, using antibodies to the Fc (fragment-crystallizable) region of the molecule. While drug target-specific strategies deliver better specificity, developing specific antibodies for each biologic of interest takes considerable time and investment. More general approaches have recently been developed using sample enrichment steps such as immunoaffinity enrichment using magnetic beads, combined with semi-targeted capture for successful sample extraction.5 These enrichment steps concentrate the target drug molecule and help to lower the complexity of the sample background.
Sample preparation for LC-MS-MS-based quantification of large molecules can involve extensive workflows, especially when dealing with proteins or peptides above 10 kDa in molecular weight. These are not always suitable for direct MS-MS analysis, so quantification must be performed on just a small portion of the protein, using a “signature” peptide that has a unique m/z ratio to provide a representative concentration for the intact protein.2 The drug target therefore has to undergo proteolytic digestion and quantification of multiple unique signature peptides; this process is widely accepted but is not always straightforward to incorporate into GLP laboratory workflows.
Furthermore, digestion of the target protein may cause variable peptide release, which can have a detrimental impact on the accuracy of signature peptide quantification data. These sample preparation steps therefore need to be highly optimized with a robust and proven protocol to achieve reproducible data with lower limits of quantitation (LLOQ) in the low ng/mL range.6 Such optimization typically involves enrichment and semi-purification of the analyte, and may also require controlling digestion of the target protein to avoid irregular signature peptide release.4 These procedures can be lengthy and complex, and knowing which sample preparation and clean-up procedures to use for different peptides requires substantial expert knowledge. Recently, there has been increasing interest in combining LBA immunoaffinity enrichment with LC-MS-MS quantification to combine the greater specificity and broader immunocapture capabilities of LBAs with the sensitivity and selectivity of LC-MS-MS technologies.5 This should result in the wider availability of pre-optimized published methods that will assist biopharmaceutical laboratories in developing sample preparation more efficiently for robust and stringent data.
There has been increasing demand for automated sample preparation technologies to increase the precision and accuracy of assays, as well as save time and expense. Automation of many laboratory procedures has become commonplace over the past 20 years, and the particularly laborious analytics needs of biologics is driving further demand for automation within LC-MS-MS workflows.
A number of front-end autosamplers and platforms have become available that automate sample preparation. More recently, instrumentation providers have begun developing complete workflow-orientated solutions that provide the hardware and software required for LC-MS-MS with biologics.
Improving Sensitivity of Detection
LC-MS-MS-based bioanalysis of biologics requires excellent MS-MS sensitivity and selectivity to quantify proteins and peptides accurately in the low ng/mL range. Quantification issues that may arise often relate to the polarity or adsorptive properties of peptides, as well as interference from background proteins, which can reduce both sensitivity and selectivity.2 Non-specific binding, poorly soluble, and poorly fragmenting peptides can also affect MS-MS quantitation and multiple reaction monitoring (MRM), all of which affect data accuracy.
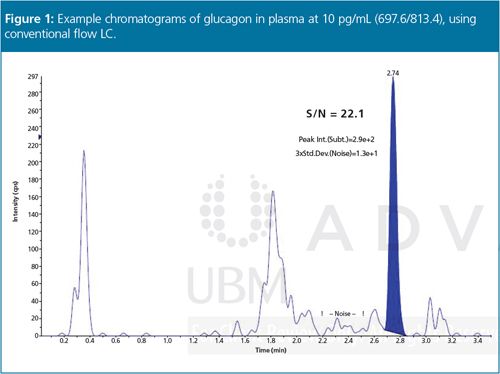
Several LC-MS-MS technology advances have recently been made that can help to overcome many of these problems. In particular, the enhancement of ionization efficiency and ion transmission in the latest triple quadrupole instruments have significantly improved sensitivity, making it possible to detect biologics at picogram levels (Figure 1) or even sub-femtogram levels.7 Technology advances within these instruments include better collisional focusing of ions, which brings more ions to the detector, as well as improvements to the detector’s dynamic range, to enhance sensitivity and performance for bioanalysis.
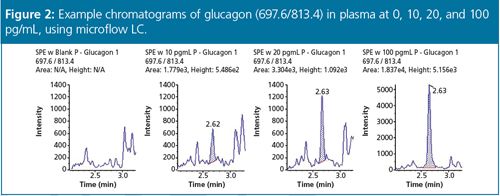
They can also be combined with microflow LC systems, allowing reduced injection volumes to be used when samples are limited, while still obtaining robust sensitivity at low picogram levels7 (Figure 2). Another advantage can come from using mass spectrometers with dual (switchable) mass ranges, which allows ions of different mass to pass through the quadrupole so that both small and large molecules can be analyzed without compromising sensitivity.
Improving Selectivity During Analysis
Selectivity can still be problematic when detecting peptides that are expressed at low levels, making it difficult to separate these out from background proteins. Methods have been developed to improve selectivity by providing an additional degree of separation, either after ions enter the mass spectrometer or after MS-MS selection. One approach is to use MRM3 scanning techniques to improve MRM peak shapes and signal-to-noise ratios. MRM3 can be performed using hybrid triple quadrupole mass spectrometers that have a linear ion trap for additional fragmentation of the primary product ions. Similarly to standard MRM, precursor ions are filtered then fragmented and filtered again in the third quadrupole. In MRM3, however, these second filtered precursor ions are fragmented and scanned again for improved selectivity.8-10 This approach means that overlapping or competing ions are filtered out, resulting in improved quantitation of the secondary product ions and lower limits of quantitation, which is advantageous for bioanalysis of biologics.
Alternatively, high resolution triple quadrupole time-of-flight (TOF) mass spectrometers enable the use of so-called MRMHR workflows,11,12 where all precursor ion fragments are detected by the TOF analyzer at high resolution and high mass accuracy. During MRMHR, ions are extracted post-acquisition at narrower extraction widths than are typically used with triple quadrupole-based experiments. This allows more accurate and specific analyte detection, even in complex biological matrices.2
Another approach is to use differential ion mobility separation techniques, where the ions are separated on mobility differences (that is, in trajectory rather than in time) as they move between a set of planar plates with high and low energy fields applied.13 This differential mobility spectrometry removes background components by providing orthogonal separation between the LC and MS stages, and results in a highly robust and fast technique with short MRM cycle times.13
Conclusions
LBAs are widely used for bioanalysis of small and large molecules, but these assays can be lengthy to develop and have limited selectivity and sensitivity. LC-MS-MS-based bioanalysis can offer important advantages over immunoassay-based techniques, but LC-MS-MS with large molecules introduces several challenges affecting sensitivity, selectivity, and data analysis. By combining traditional LBA immunoaffinity assays with the latest LC-MS-MS technologies, bioanalysts can achieve efficient immunocapture and sample extraction of large molecules, followed by highly sensitive and selective data analysis. Significantly improved selectivity can also be achieved in biological samples that have numerous highly abundant proteins by using MRM3 and differential ion mobility separation technology. Further advances in software and automation are being made to increase throughput, and enhance method development and optimization.
Although LC-MS-MS technologies have certainly advanced to be more suitable for biologics bioanalysis, the array of mass spectrometry technologies and techniques, sample preparation methods and reagents could be daunting for non-experts needing to develop a new biologics bioanalysis workflow. Accordingly, some instrumentation vendors offer integrated packages for biologics analysis that include sample preparation kits, separation and mass spectrometry technologies, and standardized software.
These developments are making LC-MS-MS more accessible for biologics bioanalysis, helping scientists to accelerate their large molecule drug development. The latest instrumentation and software developments will deliver significant improvements in the quality and reliability of bioanalysis studies, delivering more robust and compliant data that have important implications for patient safety.
References
- L.M. Jarvis, Chemistry & Engineering News93(5), 11-16 (2015).
- L. Baker and S. Ramagiri, in Biologics Bioanalysis - Guide to Innovation, pp. 6-29 (Sciex technical series) (2014).
- G. Hopfgartner, A. Lesur, and E. Varesio, Trends in Analytical Chemistry48, 52-61 (2013).
- I. van den Broek, W.M.A. Niessen, and W.D. van Dongen, Journal of Chromatography B929, 161-179 (2013).
- I. Moore, C. Yang, G. Impey, W. Woroniecki, L. Xiong, H-F. Liu, L. Baker, and S. Ramagiri, Sciex Application Note, publication no. RUO-MKT-02-2258 (2015).
- R. Bischoff, K. Bronsema, and N.C. van de Merbel, Trends in Analytical Chemistry48, 41-51 (2013).
- Sciex iMethods for Pharma and BioPharma (2015). Publication no. 8540113-01.
- S. Ramagiri, L. Olson, I. Impey, and C. Fernandez-Metzler, Biologics Bioanalysis - Guide to Innovation, pp. 86-89 (Sciex technical series) (2014).
- Y. Xu, J.P. Gutierrez, T-S Lu, H. Ding, K. Piening, E. Goodin, X. Chen, K. Miller, and Y-X Li, Biologics Bioanalysis - Guide to Innovation, pp. 78-81 (Sciex technical series) (2014).
- Y.T. Fortin, A. Salvador, J.P. Charrier, C. Lenz, F. Bettsworth, X. Lacoux, G. Choquet-Kastylevsky, and J. Lemoine, Biologics Bioanalysis - Guide to Innovation, pp. 82-85 (Sciex technical series) (2014).
- B. Simons, Sciex Application Note, publication no. 0450110-01 (2010).
- K. Lewis-Torpey, X. Wang, and C. Hunter, Sciex Application Note, publication no. 4040211-01 (2011).
- S. Taylor, The Column10(15), 14-17 (2014).
Suma Ramagiri received her BSc in biochemistry and MSc in organic chemistry in India. She did her PhD in analytical chemistry and post-doctoral research at the University of Tennessee Health Sciences, Memphis, Tennessee, USA. She has more than 12 years of experience in the analytical instrumentation field from working at Dr Reddy’s Laboratories Hyderabad, India, GTx. Inc., and ED Labs in Memphis. She has worked on many drug discovery and development projects for anti-bacterial, anti-inflammatory, and anti-cancer drugs, and DMPK studies and GLP bioanalysis. In her current role as a Global Application Manager, Pharmaceutical & CRO business at Sciex, Suma leads analytical and bioanalytical workflows in both traditional small molecule and also various biologic compound classes such as peptides, proteins, oligonucleotides, monoclonal antibodies, and antibody drug conjugates, a growing segment within pharma and biopharma. She drives worldwide efforts in application development and analytical product development to enhance bioanalytical quantitation, drug metabolism, biomarker quantitation, metabolomics, and impurity analysis. Suma has over 15 publications in scientific journals, presented at conferences and trade shows worldwide.

New Study Reviews Chromatography Methods for Flavonoid Analysis
April 21st 2025Flavonoids are widely used metabolites that carry out various functions in different industries, such as food and cosmetics. Detecting, separating, and quantifying them in fruit species can be a complicated process.
Extracting Estrogenic Hormones Using Rotating Disk and Modified Clays
April 14th 2025University of Caldas and University of Chile researchers extracted estrogenic hormones from wastewater samples using rotating disk sorption extraction. After extraction, the concentrated analytes were measured using liquid chromatography coupled with photodiode array detection (HPLC-PDA).

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)