Standardizing Proteomics
LCGC North America
Following the sequencing of the human genome, the biological science community has moved to tackle the human proteome.
As establishment of the sequence of the human genome neared completion, it was recognized that the next task for the biological science community would be to characterize the working products of the genome, which are largely proteins. The emerging field of proteomics set as its goal the identification and measurement of all the proteins in a cell or tissue with the hope that, in so doing, candidate proteins for disease biomarkers or drug targets would be found. This is proving to be a daunting task, given that each of the approximately 25,000 genes can give rise to multiple protein products through splicing and introduction of posttranslational modifications. An added complication is the huge range of protein concentrations — usually many orders of magnitude — and the possibility that the most interesting ones are in low abundance.
A host of technologies and experimental approaches has been applied to address the proteomics problem. The most widely used is the "bottom-up" approach. Proteins in an extract or lysate are digested with a proteolytic enzyme (typically trypsin), separated by reversed-phase liquid chromatography (LC), and introduced online into the electrospray ionization source of a mass spectrometer. Peptide ions are resolved in a precursor ion scan, and several of the most intense ions in the scan are subjected to fragmentation, a process referred to as LC–tandem mass spectrometry (MS) or LC–MS-MS. The combination of parent ion mass values and the fragment ion masses are submitted to a search engine, which matches the peptide and fragment masses with entries in a protein database to produce a protein identification. Quantitative information on one, several, or all proteins can be obtained by label-free methods such as spectral counting or peak intensity measurements, by introduction of stable, isotopically labeled tags, or by inclusion of a heavy-labeled version of one or several peptides found in the protein ("proteotypical peptides") (1). In the case of complex samples such as body fluids or cell lysates, the number of potential analytes can be enormous. A cell typically expresses several thousand proteins in any given state, each protein can generate up to dozens of peptides, and each peptide can appear in several charge states in the mass spectrum. Therefore, a single proteomic sample could conceivably contain upwards of 500,000 species or more. To reduce the complexity of the analytical problem, samples often are subjected to prefractionation before LC–MS using techniques such as 1D or 2D gel electrophoresis (2), in-solution isoelectric focusing (3), or multidimensional high performance liquid chromatography (HPLC) (4).

Tim Wehr
There are several limitations to the bottom-up approach when applied to complex samples. First, because of the wide dynamic range in protein concentrations, and the inability of a mass spectrometer to sample all peptide ions, the approach is inherently biased toward higher abundance proteins. Second, the large number of peptides in a complex digest combined with a limited number of MS duty cycles in an analysis reduces sampling reproducibility for low-abundance peptides. Third, information about peptide isoforms (for example, posttranslational modifications) can be lost. Finally, the large number of variables in the bottom-up experiment can compromise the reproducibility in intralaboratory and interlaboratory analyses. These are summarized in Table I.
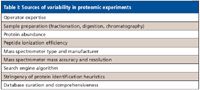
Table I: Sources of variability in proteomic experiments
Because of the large number of variables and lack of expertise in many laboratories, the quality of the data in early proteomics studies was poor and the field developed a tarnished reputation due to poor reproducibility. Over the last several years, efforts to standardize proteomics protocols and to provide reference samples have been mounted by several organizations. Five of these organizations are listed in Table II with their URLs. This installment of "Directions in Discovery" will review each of these organizations and programs they have initiated.

Table II: Organizations for standardizing proteomics
The ABRF Proteomics Research Groups
The Association of Biomolecular Research Facilities (ABRF) is a membership organization comprised of approximately 1000 members representing over 250 core laboratories in government, academia, research and industry. The organization supports 13 active several research groups created for assessment of technologies used in core facilities. Three of these focus on proteomics technologies, the Proteomics Research Group (PRG), the Proteomics Standards Research Group (sPRG), and the Proteome Informatics Research Group (iPRG).
The Proteomics Research Group: The goal of the PRG is to foster awareness and education about modern methods of protein analysis. Its primary activity is to sponsor annual research studies that examine current techniques and capabilities. Each year a specific analytical question in proteomics is addressed by a sample analysis program open to all ABRF members and outside investigators on a first-come-first-served basis. Samples typically are distributed in late fall, and results submitted anonymously to the PRG in time for them to be collated and presented in a report at the annual ABRF meeting in February–March each year. Preparation of the samples is organized by the members of the PRG and analyzed by experienced member laboratories to confirm sample quality. A list of samples and the associated analytical task for PRG studies over the last four years is presented in Table III.

Table III: PRG collaborative sample analysis programs for 2006â2009
The Proteomics Standards Research Group: The Proteomics Standards Research Group was organized to promote and support the development and use of proteomics standards to permit ABRF members and member laboratories to evaluate their ability to produce predictable and consistent results. Technical standards include reference materials, data sets, conditions and procedures. Like the PRG, the sPRG distributes samples each year to ABRF laboratories and outside investigators for collaborative studies. A list of study topics, samples, and analytical tasks for the last three collaborative studies is presented in Table IV.

Table IV: sPRG collaborative sample analysis programs
The Proteomics Informatics Research Group
The mission of the Proteomics Informatics Research Group is to educate proteomics research laboratories on the best application and practice of bioinformatics toward accurate and comprehensive analysis of proteomics data. The iPRG actively supports and participates in the development and advancement of new algorithms, software tools, and strategies for proteome informatics with the goal of both educating and introducing these technologies to the membership. The iPRG has sponsored two collaborative studies, the first in 2008 and the second in 2009. The first study evaluated the ability of participants to determinate identities of proteins in a complex mixture of proteins present in a single mass spectral dataset. Specifically, the study allowed participating laboratories to evaluate the use of bioinformatic tools to make and consolidate protein identifications from a common data set and a common reference database. The 2009 study evaluated the capability of participating laboratories in determining significantly different proteins between two complex samples. Datasets representing five technical replicates of each sample were provided to allow participants to determine reproducibility of the differences.
The Biological Reference Material Initiative
The Biological Reference Material Initiative (BRMI) is a collaboration between the National Institute for Biological Standards and Control (NIBSC) and the Proteomics Facility of the Imperial College in London. The three-year program will establish reference materials from human organs, tissues, cells, and body fluids for the standardization of proteomics methods and for training. The initial objective will be development of reference materials for 2D gel electrophoresis. In the first phase, a plasma reference material (PRM) will be prepared from WHO international standards of freeze-dried human plasma stocked at NIBSC. Reproducibility of PRM analysis between NIBSC and Imperial College will first be assessed, then the material will be made available to other proteomics laboratories. The BRMI will work in collaboration with the HUPO Protein Standards Initiative for data collection and analysis. The second phase of the BRMI will expand to other reference materials that will be cell–tissue–organ specific.
Clinical Proteomic Technology Assessment for Cancer
CPTAC is one of three major components of the Clinical Proteomics Technologies Initiative for Cancer (CPTI), a five-year program funded by the National Cancer Institute in 2006. The goal of the CTPAC initiative is to build the foundation of technologies, data, reagents, reference materials, and analysis systems needed to advance protein biology for cancer diagnosis, treatment, and prevention. CTPAC established a collaborative network of five teams, located at the institutions listed in Table V. The five multidisciplinary teams were selected based upon their expertise in proteomics and proteomics technologies. The objectives of these teams are described in Table VI. Since its inception, CPTAC has created eight working groups to address specific concerns in proteomics (Table VII). Of these, one of the most active has been the Unbiased Discovery working group, which has worked to provide metrics for troubleshooting, for documenting system performance, and for establishing system suitability. An initial study of a 20-protein mixture distributed by NIST revealed wide differences among the participating laboratories, and efforts were launched to determine the sources of variability. Subsequently, the yeast proteome was selected as a complex sample for assessing variability and to develop standard operating procedures (SOPs), protocols, and strategies to reduce variability. Linear ion trap mass spectrometers (LTQs, Thermo Fisher Scientific, Waltham, Massachusetts) or LTQ Orbitrap systems were selected as the basis for the LC–MS platforms. A family of 44 system performance metrics was adopted to monitor individual elements of the platform. These can be grouped under the categories of chromatographic performance, MS1 (precursor) signal, MS2 (fragment) signal, dynamic sampling, and peptide identification (5). Interlaboratory variation in these metrics indicated which system components were most problematical for standardization. Intralaboratory monitoring of these metrics enable preparation of "instrument health charts" to recognize when performance is drifting out of control (6). Use of such metrics to characterize LC–MS-MS performance will provide a basis for laboratories to benchmark performance, improve methodology, and evaluate new technologies.

Table V: CPTAC institutional teams
The CPTAC Targeted Verification working group is pursuing studies to evaluate the performance of protein quantitation by liquid chromatography–multiple reaction monitoring mass spectrometry (LC–MRM-MS) in plasma (7). These interlaboratory studies are identifying sources of variability and performance characteristics of LC–MRM-MS assays for peptides using Applied Biosystems (Foster City, California) Q-Trap and Thermo Quantum instruments.

Table VI: CPTAC objectives
The Fixing Proteomics Campaign
Fixing Proteomics is a web-based, noncommercial, technique-independent campaign dedicated to solving the experimental challenges that have hindered proteomics from delivering on its potential. The campaign is built around the website (www.fixingproteomics.org) and is being developed by voluntary contributions from key people in the field. The campaign was organized by a group of 10 founders, three in industry and seven in academic research institutions. The website is oriented strongly toward 2D gel electrophoresis.
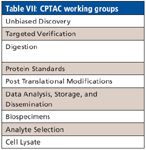
Table VII: CPTAC working groups
Fixing Proteomics provides practical advice on how to achieve cross-laboratory reproducibility including a four-step approach to Fixing Proteomics and protocols from the HUPO Reproducibility Study. The four-step approach encompasses experimental design, execution of pilot experiments with multiple biological replicates, confirmation of the results of within-laboratory pilot experiments, and confirmation of cross-laboratory reproducibility. Detailed discussion of the four steps includes presentations on issues in data analysis and design of quantitative proteomics experiments. Also included is information on two commercially available proteomics standards, the Universal Proteomics Standard for Protein Mass Spectrometry (from Sigma Aldrich, St. Louis, Missouri) and the HUPO Gold MS Protein Standard (developed by Invitrogen, Carlsbad, California).
A feature of the Fixing Proteomics site is the presentation of detailed protocols for proteomics experiments. These include a protocol for analysis of protein complexes by 1D polyacrylamide gel electrophoresis (PAGE) and tandem MS, a protocol for cell and tissue preparation for 2D PAGE, a general protocol for 2D PAGE, and a protocol for 2D gel analysis of a HeLa cell standard being developed at the Novartis Institutes for Biomedical Research. The site also presents two reports of a 2D gel reproducibility study in which a paired set of H. influenzae lysates was analyzed by five laboratories. The site also provides links to several publications, including research papers, posters presented at national and international meetings, and reports related to the Fixing Proteomics campaign in journals such as Science and Nature.
The Fixing Proteomics site is designed to be interactive, and visitors to the site are invited to provide feedback and contribute information about problems in proteomics and possible solutions.
The Human Proteome Organization
The international Human Proteome Organization (HUPO) was founded in 2002 with the intent of building a framework for the application of proteomics to the solution of biomedical problems. The organization is structured in 11 separate initiatives, with each initiative based in one country and focusing on a specific organ or aspect of proteome research (Table VIII). The Proteomics Standards Initiative (PSI) is tasked with defining standards for data representation in proteomics to facilitate data comparison, exchange, and verification. The Minimum Information About a Proteomics Experiment (MIAPE) defines guidelines for adequately reporting a proteomics experiment, providing requirements for journals and data repositories. The PSI currently is organized in six working groups, with each working group focused on a particular proteomics technology (Table IX).

Table VIII: HUPO-sponsored scientific initiatives
While the PSI focuses on documentation, the objectives of the HUPO New Technology and Resources Committee are training and education. This organization, which operates independently from the Initiatives structure, is developing test mixtures for collaborative studies among multiple laboratories. The first of these, a qualitative study of protein identification, has been completed and has been submitted for publication. The second phase of the program will be the development of a test mixture to assess quantitative methodology.
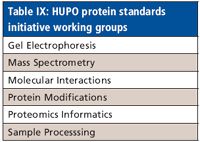
Table IX: HUPO protein standards initiative working groups
Conclusion
As proteomics emerged as a concentrated field of study early in this decade, concerns arose about the quality and reproducibility of proteomic data. The intervening years have seen dramatic improvements in the performance and robustness of MS instrumentation. This has been accompanied by improvements in sample preparation and fractionation techniques and in bioinformatics software. Going hand-in-hand with improvements in proteomics tools has been the increased attention given to standardizing experimental procedures and to providing test samples that allow proteomics laboratories to assess and improve their expertise. The five organizations reviewed here all have standardization programs in place that will be necessary for the success of proteomics.
Tim Wehr "Directions in Discovery" editor Tim Wehr is staff scientist at Bio-Rad Laboratories, Hercules, California. Direct correspondence about this column to "Directions in Discovery," LCGC, Woodbridge Corporate Plaza, 485 Route 1 South, Building F, First Floor, Iselin, NJ 08830, e-mail lcgcedit@lcgcmag.com.
References
(1) T. Wehr, LCGC 25(10), 1030–1040 (2007).
(2) T. Wehr, LCGC 19(7), 702–711 (2001).
(3) T. Wehr, LCGC 26(9), 930–936 (2008).
(4) T. Wehr, LCGC 20(10), 954–962 (2002).
(5) D.L. Liebler, "Performance and Optimization of LC- MS/MS Platforms for Proteomic Analysis: Interlaboratory studies," presented at the US HUPO meeting, San Diego, California (2009).
(6) S. Stein, L. Kilpatrick, D. Tcchekhovskoi, P. Rudnick, and E. Yan, "Measuring Variability in Shotgun Proteomics Experiments," presented at the 56th ASMS conference, Denver, Colorado (2008).
(7) S.C. Hall, "Reproducibility of Protein MRM-Based Assays: Towards Verification of Candidate Biomarkers in Human Plasma," presented at the US HUPO meeting, San Diego, California (2009).


.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)











