Single-Cell MS and High-Spatial-Resolution MS Imaging Under Ambient Conditions Using a Novel Sampling Device
LCGC North America
We have developed a miniaturized, multifunctional device, called the "Single-Probe," that is capable of probing small targets and of sampling and ionizing molecular species.
We have developed a miniaturized, multifunctional device, called the "Single-Probe," that is capable of probing small targets and of sampling and ionizing molecular species. It has been coupled with a mass spectrometer and applied to a variety of applications, including single-cell MS and MS imaging of biological tissues, and detection of sulfated proteins. In this article, we describe the probe and summarize recent work applying it to single-cell MS and MS imaging studies.
Mass spectrometry (MS) is a crucial analytical technique based on the measurement of mass-to-charge ratios (m/z) of ions of interest. Traditionally, MS has been used to analyze gas-phase samples or samples in liquid solution. More recently, combined with advances in sampling and ionization methods, instrument automation, and data processing, MS has been used to analyze a broad range of samples. Of particular interest is the application of MS to emerging techniques such as single-cell MS analysis and MS imaging of biological tissues.
We developed a novel apparatus, called the "Single-Probe," which is a miniaturized, multifunctional device. The name "Single-Probe" reflects the unique features of this sampling device: integrated design, multiple functions, and versatile applications.
The single-probe device comprises several integrated, microminiaturized components, and is capable of probing extremely small targets and sampling and ionizing molecular species. The probe can be coupled with a mass spectrometer, and in this way it has been applied to various applications including single-cell MS, MS imaging of biological tissues, and the detection of sulfated proteins. In this article, we describe the probe and summarize our recent work using it in single-cell MS and MS imaging studies.
MS Analysis of Live, Single Cells
Single-cell analysis is a new field that has the potential to reshape approaches to biological and pharmaceutical bioanalytical research (1–3). The mechanisms of biological processes at the cellular level are important for understanding life. It is well known that cells of the same tissue can behave differently, even under the same conditions. Yet the current methods for analyzing the chemical makeup of bulk cellular composition (such as with cell lysates) provide limited information because the measured results are an average of all of the samples combined. In particular, any cell preparation method is likely to alter a cell's environment and structure, changing the chemical composition of cellular content. MS has been used for single-cell analysis because of its high sensitivity-it is capable of detecting a broad range of molecular species. Single-cell MS constitutes a new field in bioanalysis, and a variety of techniques to apply it have been developed. The major difference among various approaches to single-cell MS lie in their sampling and ionization methods. To date, various MS methods have been developed for single-cell analysis, including secondary-ion mass spectrometry (SIMS) (4–6), matrix-assisted laser desorption–ionization (MALDI) (4,7,8), laser desorption–ionization mass spectrometry (LDI-MS) (9,10), capillary electrophoresis–electrospray ionization MS (CE–ESI-MS) (11–13), and live, single-cell MS (14–18). Although these established single-cell MS techniques have served in a variety of studies, their inherent limitations constrain their broader application. For example, SIMS provides high spatial resolution, but it must be operated under high vacuum conditions. MALDI, currently the dominant technique because of its good resolution (19), requires time-consuming sample preparation, and the required matrix changes the conditions of biological samples (20). Therefore, neither SIMS nor MALDI-MS can be used for live cells. Single-cell MS analysis of living cells has been reported using the live single-cell MS technique (14–18), in which a sharp nanoelectrospray ionization (nano-ESI) emitter penetrates the cell membrane and extracts cellular contents. The intracellular contents are mixed with the ionization solvent in the emitter and the mixture then is transferred to a mass spectrometer for analysis. Because the sampling, cell-extraction preparation, and measurement steps must be performed separately, this technique cannot be used for in situ or real-time cellular analysis.
The single-probe device, which can be coupled with a mass spectrometer, can overcome the barriers to live, single-cell analysis. The single-probe device combines two capillaries into a laser-pulled, dual-bore, quartz needle. One capillary (the solvent-providing capillary) delivers the solvent to the tip of the device; a small liquid junction is formed between two bores at the quartz needle, to pick up the component of the cell. The solution containing the cellular content is drawn by the other capillary (the nano-ESI emitter) via capillary action, and then is ionized for immediate MS analysis (Figure 1a). With a diameter of 6–10 μm, the probe tip is small enough to be inserted into an individual cell (Figure 1b). The entire single-cell MS experimental system includes an automated X,Y,Z-translational stage system, two digital light microscopes, and a Thermo LTQ Orbitrap XL mass spectrometer.
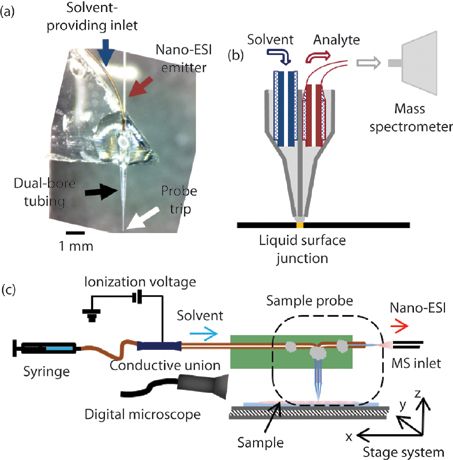
Figure 1: (a) Photograph of the constructed single-probe device, zoomed in on the region around the tip. (b) Schematic of the construction of the single-probe device, showing the solvent-providing capillary, the dual-bore tubing with the pulled probe tip, and the nano-ESI emitter. (c) Diagram showing the setup for the operation of the single-probe device for sample acquisition. (Adapted with permission from reference 37.)
During our experiment, the cell sample was placed on the stage system, which is controlled by LabView software, developed by the Laskin group (21). Cell penetration is achieved by precisely lifting the Z-stage using the microscopes as the visual guide (Figure 1c). To ionize the sampled species for MS analysis, an ionization voltage of approximately 3 kV energizes the conductive union. The voltage charge is transmitted to the nano-ESI emitter continuously through the solvent inside the single-probe device. The ionization voltage cannot be applied directly on the nano-ESI emitter, because the emitter is too small (<5 mm). The continuous flow of sampling solvent provides a consistent, fresh, liquid junction from which to extract cellular compounds for MS analysis in real-time. The single-probe MS technique allows for in situ, live analysis of a single cell under ambient conditions, with little or no prior sample preparation required.
Before the single-cell MS experiments, the live cells are rinsed with phosphate-buffered saline (PBS) to remove extracellular culture medium or drug compounds. Placed on the Z-translation stage, the cell-containing plate can be lifted precisely, by 0.1-μm increments, and monitored by means of the microscope. The penetration is confirmed visually using the appended microscope and sideways camera. The insertion of the probe is monitored by an immediate change of mass spectrum over the background signal. A time delay, of 1 to 2 s, is usually observed between the moment of insertion and detection of the MS signal. Single-probe sampling continues until no cytoplasm-MS signals are observed. Then the single-probe device is lifted, and the constant solvent flow flushes the device, preventing any carryover effects. Each cell measurement requires approximately 3 min, making this a high throughput technique. If the probe becomes clogged, a miniature coil can be placed around the clogged parts (such as the probe tip or nano-ESI emitter) and heat them for a few seconds, after which solvent flowing through the probe removes any debris. This unclogging technique increases the robustness of the single-probe device and allows its reuse. If clogging cannot be overcome, then the single-probe device can be quickly replaced with a fresh one.
We used the single-probe MS technique to successfully analyze individual HeLa cells (22) (Figure 2a) and observed cellular metabolites and lipids in the single cells (Figure 2b). In the positive mode, we identified 18 different lipid species; in the negative mode, we detected adenosine phosphates (AMP, ADP, and ATP). In the experiments of cells treated with the anticancer drugs doxorubicin, paclitaxel, or OSW-1, we detected the corresponding drugs and their metabolites in individual HeLa cells. In these experiments, HeLa cells, under normal culture conditions, were dosed with compound for 4 h at a series of concentrations: 10 nM, 100 nM, 1 μM, and 10 μM. Untreated HeLa cells served as controls. Ions for the drugs' metabolite compounds were not present in the extracellular PBS, but the compounds were detected inside the single cells.
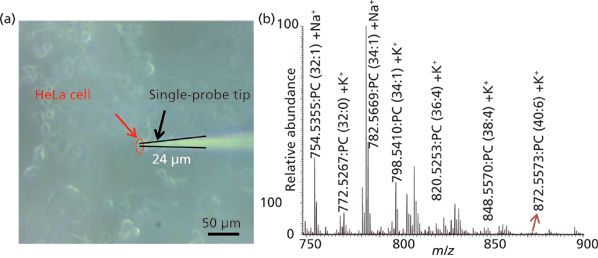
Figure 2: (a) Photograph of the tip of the single-probe device entering the target cell for acquisition of MS spectra. (b) Typical single cell analysis spectrum showing some of the identified peaks of phosphatidylcholine (PC) species in the positive ion mode (adapted with permission from reference 22).
Though still in its infancy, single-cell MS analysis has already attracted a lot of attention in the research community. Its ability to analyze samples with a level of precision previously impossible offers the potential to substantially improve our understanding of many biological systems. The single-probe device can, potentially, provide a robust tool for single-cell MS experiments. With further improvements, it will allow better and easier performance of single-cell analysis for a wide range of biological samples.
High-Resolution Ambient MS Imaging
MS imaging is an emerging technology that has attracted considerable interest from researchers in many diverse fields since its debut in 1997 (23). The process of MS imaging involves the MS investigation of the surface of a sample in a regular grid, with each individual grid position producing an entire mass spectrum. Compared with traditional MS techniques capable of analyzing the chemical composition of samples, MS imaging makes it possible to map the spatial distribution of species of interest on a sample's surface. In particular, certain MS imaging techniques can create MS images at subcellular resolution. Such images provide a wealth of information about the distribution of metabolites. These techniques can be used in various applications such as biomarker discovery (24), tissue-metabolite profiling (25), precision histological identification (such as for cancer versus normal cells) (26), and many others.
Broadly speaking, MS imaging techniques are split between two categories: nonambient and ambient. The distinction involves the way in which samples are prepared before analysis. In nonambient MS imaging, such as MALDI (27) and time-of-flight secondary-ion MS (TOF-SIMS) (28) MS imaging, the samples require the application of a matrix, the presence of a vacuum, or both to detect surface metabolites. Both of these technologies allow the imaging of samples at very high spatial resolutions. MALDI is capable of a resolution of 5 μm and TOF-SIMS at a pixel size of <1 μm. The nonambient nature of these techniques, however, can increase sample analysis time during preparation. It may also cause changes to the sample that could introduce artifacts into the resultant MS image (29).
Ambient MS imaging is a relatively recent advance. The technique involves performing MS imaging in an ambient environment in which the sample has undergone only minimal pretreatment (30). Thus, compared with nonambient methods, ambient MS imaging allows the sample-such as a tissue section from a mouse brain or a tumor-to be analyzed in an environment approaching that of its native state. The result is fewer artifacts and the potential for creating a better representation of the sample in the MS image.
The first ambient MS imaging technique, desorption electrospray ionization (DESI) MS imaging, was described in 2004 (31), and a proliferation of ambient MS imaging techniques followed (30). In DESI MS imaging, a spray of solvent is focused onto a sample and the analytes are desorbed from the sample's surface by means of a droplet-pickup mechanism. The analytes are then transmitted into the mass spectrometer for analysis. Surface-extraction techniques such as liquid microjunction surface-sampling probe (LMJ-SSP) (32), liquid-extraction surface analysis (LESA) (33), and nano-DESI (34) create a liquid-surface microjunction from which metabolites are extracted during MS imaging. The techniques have demonstrated greater sensitivity than that provided by DESI imaging (35). Nevertheless, one of the biggest obstacles for ambient MSI techniques is the lack of spatial resolution. Though DESI MSI is capable of a resolution of 35 μm (36), it is routinely performed at 100–200 μm to maintain high sensitivity throughout the analysis. Both LMJ-SSP and LESA create relatively large liquid microjunctions, with spatial resolutions exceeding 500 μm. Of the aforementioned techniques, nano-DESI MS imaging provides the greatest spatial resolution, with MS images created at approximately 12 μm in lateral resolution. Yet such performance cannot compare with the spatial resolution of nonambient techniques such as MALDI MS imaging, which are capable of imaging precision at subcellular levels.
We have applied the single-probe device to MS imaging and have been able to improve the spatial resolution of ambient MS imaging techniques. The single-probe device is readily adapted to the surface microjunction mode of MS imaging, so that a high degree of sensitivity can be achieved during the analysis (37). The probe was modified to reduce the length of the emitter, minimizing the effect of carryover and thus improving the spatial resolution of the images produced. One major advantage of the single-probe device is that its integrated design allows for relatively easy setup during MS image acquisition.
The single-probe device was used first to create MS images of standardized samples. From those images, a spatial resolution of 8 ± 2 μm was achieved using a rhodamine grid (37). The small size of the single-probe device tip (which can be as small as 6 μm) made this outcome possible. We then applied the device to the imaging of biological samples made by cryosection. That application, however, presented a much bigger challenge. Cryosectioned biological tissues are known to demonstrate a certain amount of surface heterogeneity that can introduce inconsistencies during surface microjunction formation (21). The inconsistencies would create difficulties during MS imaging that could affect the spatial resolution of the images produced. We were able to minimize this effect by creating thin sections (~12 μm) of mouse brain and kidney. In conjunction with the custom-designed LabView control software for the XYZ stage, the thinness of the tissue sections allowed the single-probe device to achieve a stable microjunction with a flat surface throughout the analysis. MS imaging of these tissues were made with a Thermo LTQ Orbitrap XL mass spectrometer, which provided high mass resolution at 60,000 m/Δm. Overall, we were able to achieve a maximum lateral resolution of 8.5 μm for an analysis of mouse kidney tissue, a resolution that is among the highest spatial resolutions reported for ambient MS imaging techniques (Figure 3). Also, the high mass accuracy made it possible to identify an entire host of metabolites, such as sphingomyelin (34:1) (725.5575 m/z, [M + Na]+), phosphatidylcholine (PC) (32:0) (734.5700 m/z, [M + H]+), PC (34:1) (782.5696 m/z, [M + Na]+), PC (36:4) (820.5256 m/z, [M + K]+), and many others with a mass confidence below 4 ppm, which allows detailed distributions of metabolites for each of the areas within the mouse kidney sample to be elucidated.
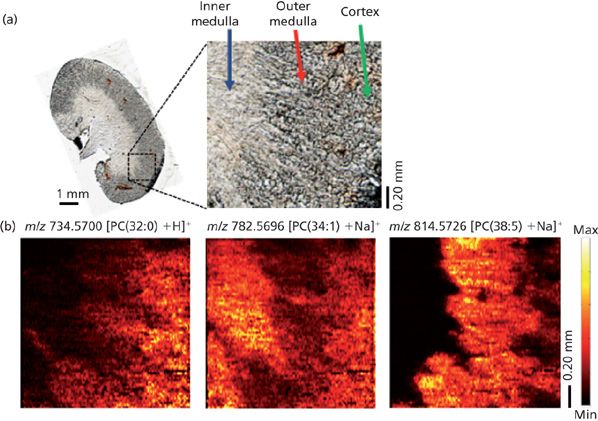
Figure 3: (a) Optical image of kidney section imaged using the single-probe device. (b) MS images of three phosphatidylcholine (PC) species showing their different distributions within the sample. The imaging resolution was at 8.5 μm. (Adapted with permission from reference 37.)
Accordingly, we expect to be able to further improve the spatial resolution of the single-probe device to the extent that it may approach the resolution offered by nonambient techniques such as MALDI MS imaging. The enhanced resolution would allow subcellular levels of MS imaging to be conducted in the ambient environment, improving analysis time as well as reducing the possibility of introducing artifacts during sample preparation. We would also like to further improve the manufacturing process for the probe so that it can become more consistent and robust in operation and user-friendly, allowing the device to be more readily used by a larger number of researchers. The single-probe device has the potential to become an effective tool for elucidating analyte distributions with high-spatial-resolution ambient MS imaging. We would like to apply this technique for investigation of an increasingly sophisticated range of applications-imaging drug distributions within tissue and biomarker discovery for cancers, to name but two-that may be of greater interest to the biological, pharmaceutical, and medical research communities.
References
(1) H. Andersson and A. van den Berg, Curr. Opin. Biotech. 15, 44–49 (2004).
(2) D.J. Wang and S. Bodovitz, Trends Biotechnol.28, 281–290 (2010).
(3) S.S. Rubakhin, E.V. Romanova, P. Nemes, and J.V. Sweedler, Nature Methods8, S20–S29 (2011).
(4) M.L. Pacholski and N. Winograd, Chem. Rev.99, 2977–3006 (1999).
(5) K. Chughtai and R.M.A. Heeren, Chem. Rev. 110, 3237–3277 (2010).
(6) E.J. Lanni, S.S. Rubakhin, and J.V. Sweedler, J. Proteomics75, 5036–5051 (2012).
(7) K.A.Z. Berry, J.A. Hankin, R.M. Barkley, J.M. Spraggins, R.M. Caprioli, and R.C. Murphy, Chem. Rev.111, 6491–6512 (2011).
(8) A. Amantonico, J.Y. Oh, J. Sobek, M. Heinemann, and R. Zenobi, Angew Chem. Int. Edit.47, 5382–5385 (2008).
(9) M.P. Greving, G.J. Patti, and G. Siuzdak, Anal. Chem.83, 2–7 (2011).
(10) P.L. Urban, T. Schmid, A. Amantonico, and R. Zenobi, Anal. Chem.83, 1843–1849 (2011).
(11) J.S. Mellors, K. Jorabchi, L.M. Smith, and J.M. Ramsey, Anal. Chem.82, 967–973 (2010).
(12) P. Nemes, A.M. Knolhoff, S.S. Rubakhin, and J.V. Sweedler, Anal. Chem.83, 6810–6817 (2011).
(13) P. Nemes, S.S. Rubakhin, J.T. Aerts, and J.V. Sweedler, Nat. Protoc.8, 783–799 (2013).
(14) Y. Fukano, N. Tsuyama, H. Mizuno, S. Date, M. Takano, and T. Masujima, Nanomedicine-UK7, 1365–1374 (2012).
(15) T. Masujima, Anal. Sci.25, 953–960 (2009).
(16) H. Mizuno, N. Tsuyama, T. Harada, and T. Masujima, J. Mass Spectrom.43, 1692–1700 (2008).
(17) N. Tsuyama, H. Mizuno, E. Tokunaga, and T. Masujima, Anal. Sci.24, 559–561 (2008).
(18) M.L. Tejedor, H. Mizuno, N. Tsuyama, T. Harada, and T. Masujima, Anal. Chem.84, 5221–5228 (2012).
(19) A. Zavalin, J.H. Yang, A. Haase, A. Holle, and R. Caprioli, J. Am. Soc. Mass Spectrom.25, 1079–1082 (2014).
(20) N. Tsuyama, H. Mizuno, and T. Masujima, Anal. Sci.27, 163–170 (2011).
(21) I. Lanekoff, B.S. Heath, A. Liyu, M. Thomas, J.P. Carson, and J. Laskin, Anal. Chem.84, 8351–8356 (2012).
(22) N. Pan, W. Rao, N.R. Kothapalli, R. Liu, A.W.G. Burgett, and Z. Yang, Anal. Chem.86, 9376–9380 (2014).
(23) R.M. Caprioli, T.B. Farmer, and J. Gile, Anal. Chem.69, 4751–4760 (1997).
(24) K. Schwamborn, J. Proteomics75, 4990–4998 (2012).
(25) I. Lanekoff, M. Thomas, J.P. Carson, J.N. Smith, C. Timchalk, and J. Laskin, Anal. Chem.85, 882–889 (2013).
(26) S. Gerbig, O. Golf, J. Balog, J. Denes, Z. Baranyai, A. Zarand, E. Raso, J. Timar, and Z. Takats, Anal. Bioanal. Chem.403, 2315–2325 (2012).
(27) A. Rompp and B. Spengler, Histochem. Cell Biol.139, 759–783 (2013).
(28) J. Brison, M.A. Robinson, D.S.W. Benoit, S. Muramoto, P.S. Stayton, and D.G. Castner, Anal. Chem.85, 10869–10877 (2013).
(29) J.C. Vickerman, Analyst136, 2199–2217 (2011).
(30) C. Wu, A.L. Dill, L.S. Eberlin, R.G. Cooks, and D.R. Ifa, Mass Spectrom. Rev. 32(3), 218–243 (2012).
(31) Z. Takats, J.M. Wiseman, B. Gologan, and R.G. Cooks, Science 306, 471–473 (2004).
(32) G.J. Van Berkel, A.D. Sanchez, and J.M. Quirke, Anal. Chem.74, 6216–6223 (2002).
(33) V. Kertesz and G.J. Van Berkel, J. Mass Spectrom.45, 252–260 (2010).
(34) J. Laskin, B.S. Heath, P.J. Roach, L. Cazares, and O.J. Semmes, Anal. Chem. 84, 141–148 (2012).
(35) W. Rao, A.D. Celiz, D.J. Scurr, M.R. Alexander, and D.A. Barrett, J. Am. Soc. Mass Spectrom. 24, 1927–1936 (2013).
(36) D.I. Campbell, C.R. Ferreira, L.S. Eberlin, and R.G. Cooks, Anal. Bioanal. Chem.404, 389–398 (2012).
(37) W. Rao, N. Pan, and Z. Yang, J. Am. Soc. Mass Spectrom.26, 986–993 (2015).
Ning Pan, PhD, earned her bachelor's degree in chemistry from Shandong Normal University, in China, in 2007. In 2010 she earned a master's degree in organic chemistry from Shandong Normal University and in 2013 she earned a PhD in analytical chemistry from Shandong Normal University and Ohio University. She is currently a Postdoctoral Research Associate in Chemistry at the University of Oklahoma. Her research focuses on single-cell mass spectrometry analysis.

Ning Pan, PhD
Wei Rao, PhD, earned his bachelor's degree in genetics at the University of Cambridge, UK, in 2005. After graduation he went on to complete his PhD in proteomics under Professor R. E. Isaac in 2010 at the University of Leeds, UK. His first postdoctoral position was with Professor D.A. Barrett at the University of Nottingham in the UK, looking at ambient surface MS imaging of proteins. He is currently employed at the University of Oklahoma under Dr. Zhibo Yang, working on high spatial and mass ambient MS imaging using the single-probe device.

Wei Rao, PhD
Zhibo Yang, PhD, obtained his B.S. (1997) and M.S. (2000) degrees from the University of Science and Technology of China. In 2005, he received his PhD in Physical Chemistry under the supervision of Professor Mary T. Rodgers at Wayne State University. He has conducted postdoctoral research with Dr. Julia Laskin at Pacific Northwest National Laboratory (2005–2008) and Professor Veronica M. Bierbaum at the University of Colorado, Boulder (2008–2012). In 2012, he joined the Department of Chemistry and Biochemistry at the University of Oklahoma as an assistant professor. His current research is focused on the development and application of novel mass spectrometry techniques for bioanalysis. Dr. Yang was the recipient of the 2014 ASMS Research Award.

Zhibo Yang, PhD
Kate Yu "MS - The Practical Art" Editor Kate Yu joined Waters in Milford, Massachusetts, in 1998. She has a wealth of experience in applying LC–MS technologies to various application fields such as metabolite identification, metabolomics, quantitative bioanalysis, natural products, and environmental applications. Direct correspondence about this column to lcgcedit@lcgcmag.com

Kate Yu

Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.
Silvia Radenkovic on Building Connections in the Scientific Community
April 11th 2025In the second part of our conversation with Silvia Radenkovic, she shares insights into her involvement in scientific organizations and offers advice for young scientists looking to engage more in scientific organizations.














