Pressure–Enhanced Liquid Chromatography as a Suitable Approach to Improve Selectivity for Large Molecule Separations
This work describes the application of a pressure-enhanced liquid chromatography (PE-LC) setup to tune the separation of various large molecules comprised of nucleic acids (oligonucleotides, messenger ribonucleic acid [mRNA], and deoxyribonucleic acid [DNA]). When adding pressure as a method development parameter, it indeed becomes possible to modify retention, selectivity, and peak width. As an example, the separation of oligonucleotides having sizes comprised between 40 and 100-mer in ion-pairing reversed-phase liquid chromatography (IP-RPLC) was drastically improved by using a stepwise pressure gradient to selectively shift the retention of a peak or group of peaks. Resolution was increased from 1.5 to 11.8 when setting a rapid high pressure step (twofold increased pressure) during the run. On the other hand, it was also possible to improve the separation of erythropoietin (EPO) mRNA and related impurities under ion-exchange chromatography (IEX). However, with this biomolecule, the best separation was achieved by reducing the pressure in the system. Finally, for another sample (DNA ladder) under IEX conditions, the pressure was found to have a limited impact on the overall selectivity. As highlighted in this work, pressure is an additional parameter that can be successfully used to develop LC methods of large biomolecules.
Operating pressure in liquid chromatography (LC) can have an impact on solute retention, especially for large molecules (1–8). Indeed, the Gibbs free energy model suggests that pressure effects are related to the change of partial molar volume when a solute is transferred from one phase of the system to the other (that is, the difference between molar volumes of the solute when adsorbed and desorbed) (9), and typically, such effects are more pronounced for large molecules. In addition to the change of molar volume, many other phenomena of a phase system may change with pressure, such as solvation, solubility, molecular conformation, pKa of the functional groups (both the stationary phase and solute), or variations in the energy of molecular interactions. All of these effects can lead to altered retention and selectivity (9,10).
Even though pressure can be a potential variable to tune the selectivity of a LC separation, it has not yet been considered as a method development parameter. For a conventional LC separation, the overall pressure (column head pressure) is constant under isocratic elution mode (pressure drops linearly across the column from the highest value at the column head to atmospheric pressure at the column exit). The head pressure might vary with composition-programmed gradient elution because of the viscosity differences of mixed solvent components (suggesting that the pressure drop along the column will follow a nonlinear trend).
To evaluate pressure effects in a controlled manner, we recently introduced a new form of pressurized separations called pressure-enhanced–LC (PE-LC) (11). An apparatus for the facile manipulation of column pressure was assembled, consisting of a two-pump system and post-column flow restriction. Such a system enables combining mobile phase and pressure gradients in the same analytical run, or to work at any constant pressure independent of the flow delivered through the column (11). The PE-LC approach was already applied to improve peptides and protein separations, resulting in previously unseen selectivities.
In this work, the PE-LC setup was utilized to improve the separation of relatively short oligonucleotides in ion-pairing reversed-phase (IP-RP) chromatography, and larger nucleic acids in ion-exchange (IEX) chromatography. We believe that these new applications will convince the readers of the high potential of PE-LC, especially for the analysis of new modalities (that is, cell and gene therapy products, such as oligonucleotides, nucleic acids, viral vectors, and lipid nanoparticles).
Experimental
Chemicals, Samples, and Columns
High performance LC (HPLC) grade water and methanol were obtained from Fisher Scientific. Tetramethyl-ammonium chloride (TMAC), sodium chloride (NaCl), 1N sodium hydroxide (NaOH) solution, 1N hydrochloric acid (HCl) solution, tris(hydroxymethyl)aminomethane (TRIS), hexafluoroisopropanol (HFIP), and triethylamine (TEA) were purchased from Sigma-Aldrich.
Oligodeoxythymidines consisting of 40, 60, 80, and 100 nucleotides (namely, dT40, dT60, dT80, and dT100) were purchased from Integrated DNA Technologies and reconstituted in water at 100 μM. An equimolar oligodeoxythymidine mixture was prepared by mixing 100-μM aliquots and diluting the oligonucleotide products to 5 μM in water prior to IP-RPLC analysis.
EPO mRNA (length of 858 nucleotide) was purchased from TriLink Biotechnologies. The sample was diluted to 25 μg/mL in water and directly injected without further preparation.
A DNA ladder (100–1517 bp) was purchased from New England Biolabs. The sample was diluted to 0.05 mg/mL and directly injected without further manipulation.
IP-RP measurements were performed on a Acquity Premier BEH C18 (50 x 2.1 mm, 130 Å, 1.7 μm) column. IEX measurements were performed using Protein Pak High Res Q (5 μm, 50 x 2.1 mm and 100 x 4.6 mm, non-porous) columns. All columns were provided by Waters.
Modified Chromatographic System
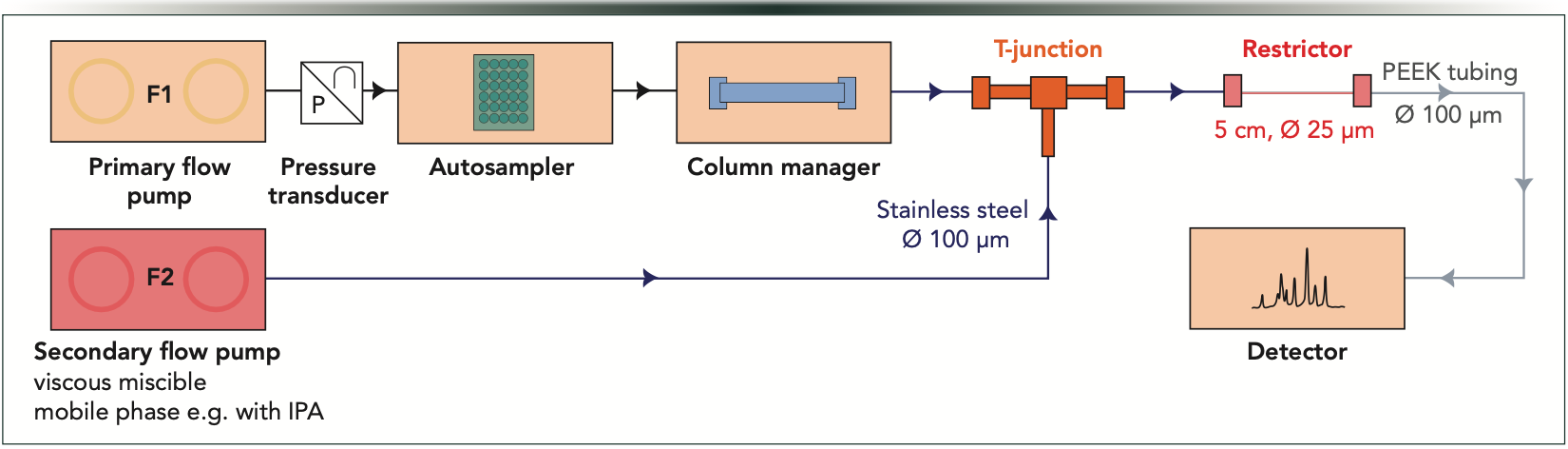
A modified ultrahigh-pressure LC (UHPLC) system with an extra secondary flow pump was used. Figure 1 shows a schematic view of the arrangement. The flow from the primary pump is directed through the autosampler and column and then to a T-junction where the primary flow meets the pressure-regulating secondary solvent flow. Then, the mixed fluid from the T-junction is directed to a short restrictor capillary tube (5 cm × 25 μm) and to the optical UV detector via polyetheretherketone (PEEK) tubing. By changing the flow rate of the secondary pump, the column will experience different pressures (independently of the flow rate set on the primary pump). The flow rate and the mobile phase entering the column can be adjusted independently of the secondary flow. With such a setup, an arbitrary pressure can be set by changing the flow rate of the secondary pump, allowing the realization of a desired pressure profile. For example, concave, convex, or any multisegmented pressure gradients can also be programmed.
Figure 1: Schematic illustration of the PE-LC system used in this work. From left to right: F1 is the primary flow rate controlled by the primary flow pump, F2 is the secondary flow rate controlled by the secondary flow pump, pressure transducer, autosampler, column manager and column, T-junction, restrictor, and detector.
Chromatographic Conditions
Oligodeoxythymidines were analyzed in IP-RP mode. Mobile phase A contained 14 mM of triethylamine (TEA) and 100 mM of hexafluoroisopropanol (HFIP) in water. Mobile phase B was a mix of A (50%) and of methanol (50%). Mobile phase B was also used for the secondary flow solvent. The primary flow rate was set to 0.2 mL/min, and a gradient of 35–45% B in 5 min was applied. The column temperature was set to 60 °C. Various flow rates and flow rate gradients were set for the secondary pump to generate different pressure programs.
Intact EPO mRNA was analyzed in anion-exchange (AEX) chromatographic mode. Mobile phase A was 25 mM TRIS (pH = 7.6), mobile phase B was 3 M TMAC in mobile phase A. The mobile phase composition gradient was 72–85% B in 10 min at a flow rate of 0.1 mL/min, and at ambient temperatures. Various flow rates were set for the secondary pump to generate different pressures. The secondary flow solvent was water. A small volume column was used for these experiments (Protein Pak High Res Q, 5μm, 50x2.1mm).
The components of the 100 bp DNA ladder were separated in AEX mode using a conventional column format (Protein Pak High Res Q 5 μm, 100 x 4.6 mm). Mobile phase A was 25 mM TRIS (pH = 7.9) and mobile phase B was 2 M NaCl in mobile phase A. A shallow mobile phase gradient (40–44% B in 30 min) was applied. The primary flow rate was set to 0.2 mL/min. Experiments were performed at ambient temperatures, and the secondary flow solvent was mobile phase A.
Results and Discussion
General Considerations
In most RP separations, the retention of large solutes increases with pressure. As a result, a high-pressure condition will cause solute velocity to decrease compared to a low-pressure condition. It is believed that the same applies to pressure gradients. Hence, a positive pressure gradient is predicted to result in a retention increase and additional band broadening. Meanwhile, a negative pressure gradient is predicted to yield a slight peak compression and retention decrease. However, please note that retention does not necessarily increase with pressure. Indeed, a negative retention trend was already reported with a pressure increase for IEX and hydro-phobic interaction chromatography (HIC) separations of proteins (12,13).
Changing the Selectivity of Oligonucleotide Separations with PE-LC
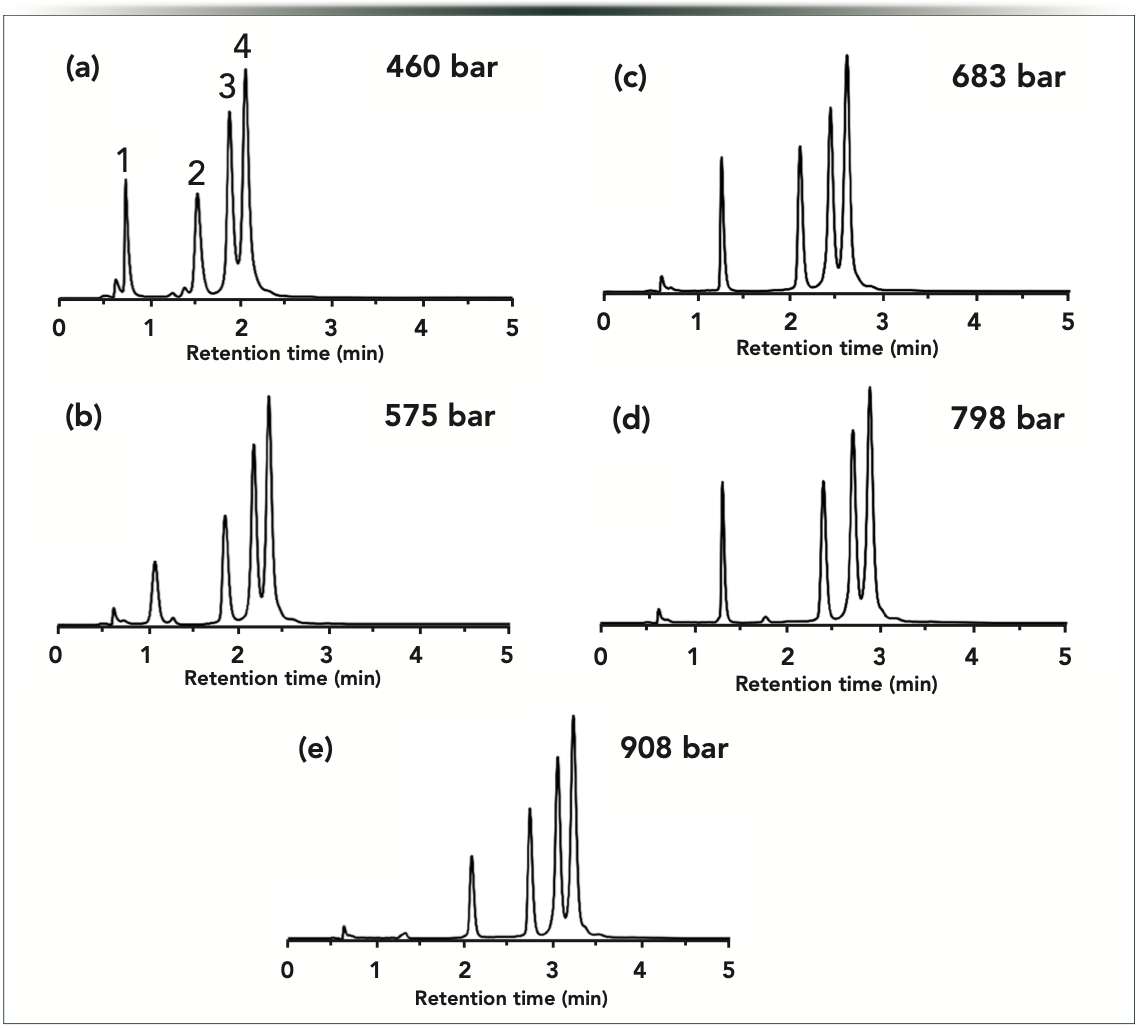
First, constant pressure experiments were performed by varying the column inlet pressure between 460 and 908 bar. Figure 2 shows the corresponding chromatograms. For these oligonucleotides analyzed in IP-RP mode, retention increased proportionally with pressure. The overall elution profile (relative peak distribution) did not change significantly, but a very minor improvement in resolution (Rs) was observed between peaks 2–3 (dT60/ dT80) and peaks 3–4 (dT80/dT100). Resolution values between peaks 2 and 3 were Rs = 2.76 at 460 bar and 3.17 at 908 bar. Although, for peaks 3 and 4, the Rs was equal to 1.20 and 1.45 at 460 and 908 bar pressure, respectively. Table I lists the obtained retention times and resolution values for the oligonucleotide separations in IP-RP mode.
Figure 2: Gradient elution separations of oligonucleotides performed at constant but different pressures. Experiments were performed at p (in bar) = (a) 460, (b) 575, (c) 683, (d) 798, and (e) 908 bar. Peaks: 1 = dT40; 2 = dT60; 3 = dT80; and 4 = dT100.
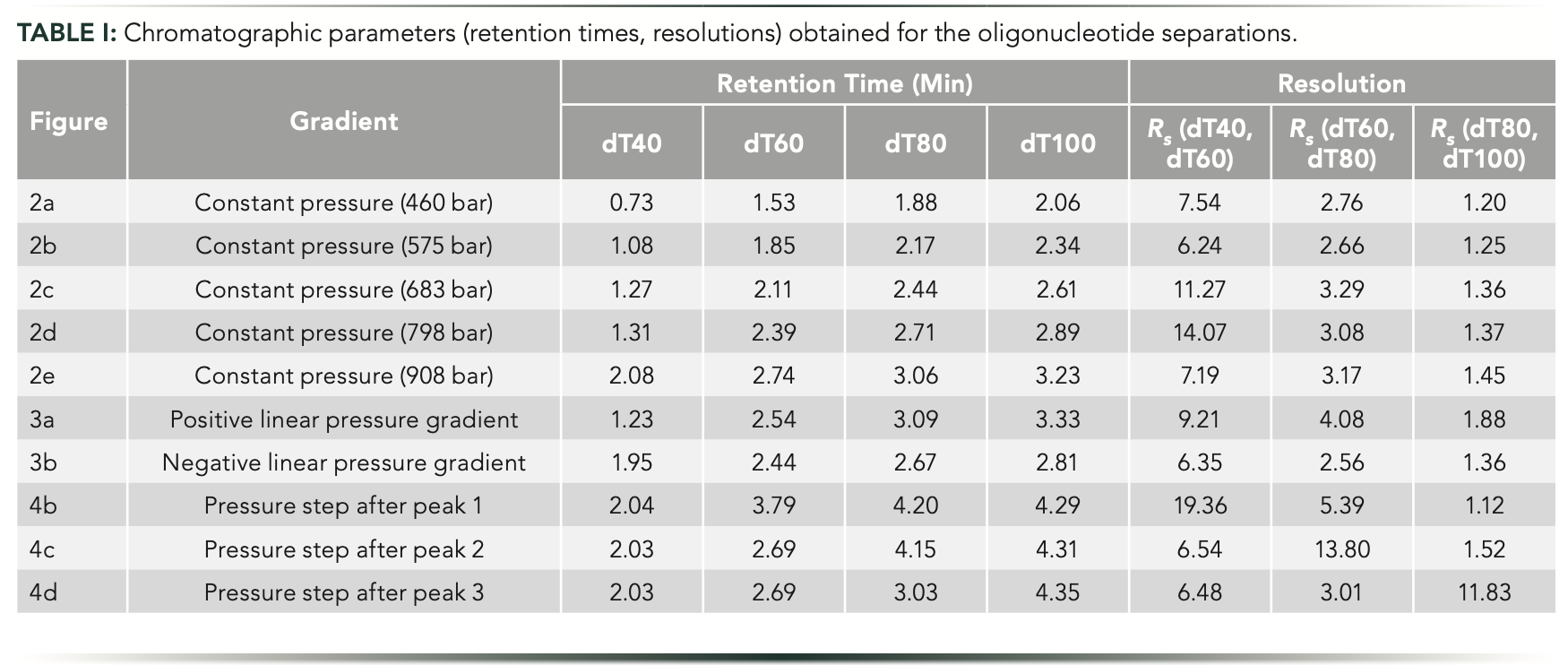
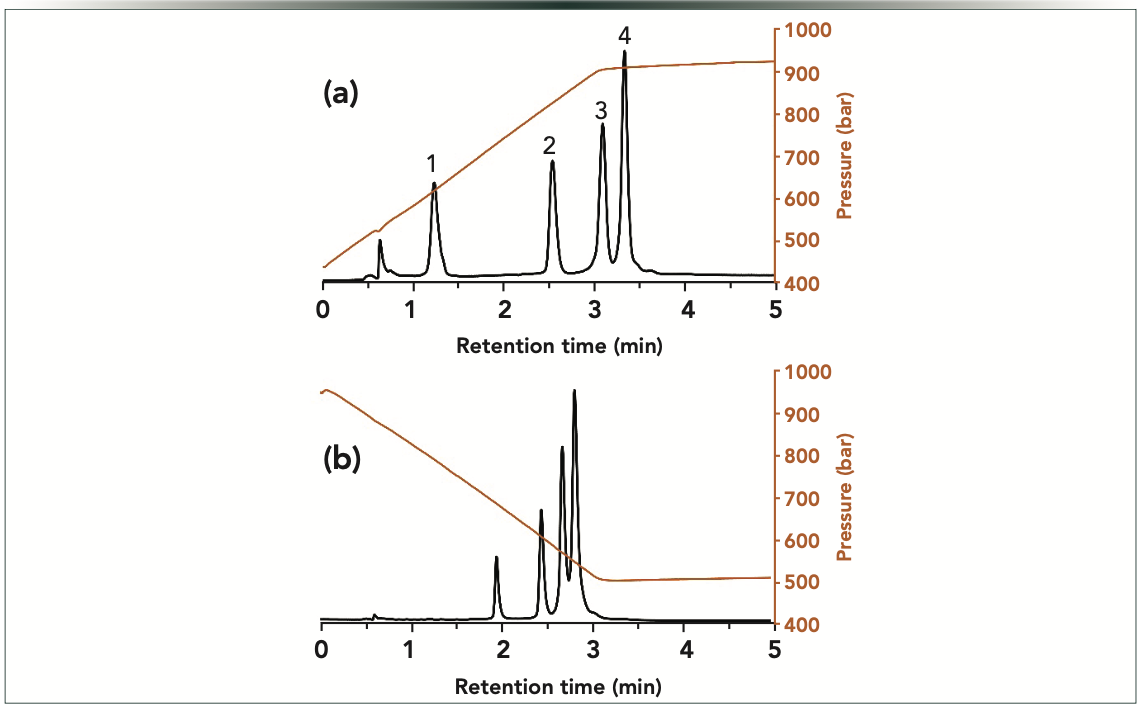
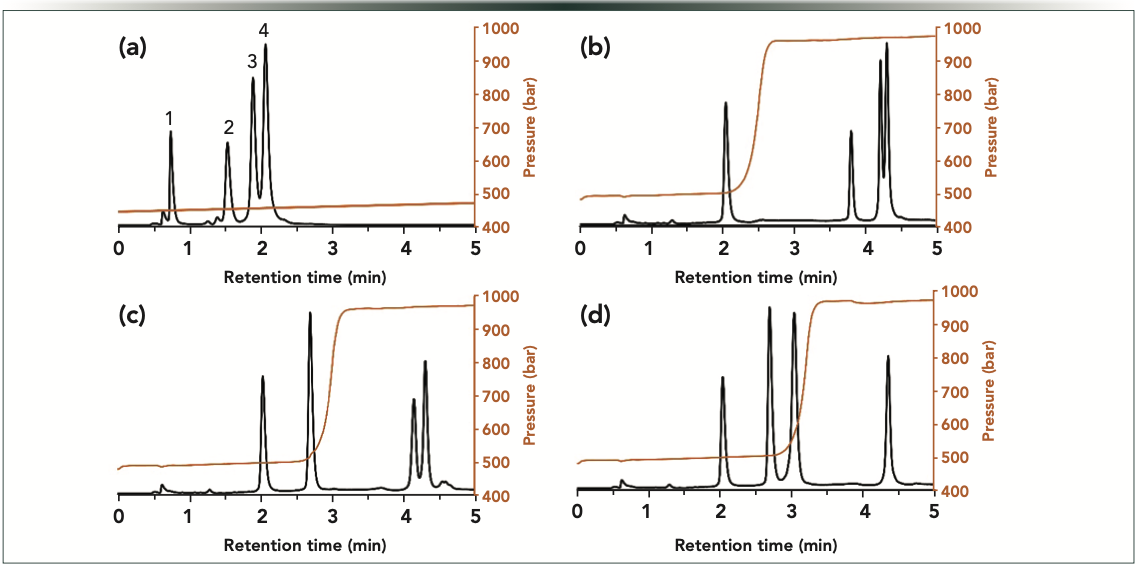
Next, we have tried to combine dual pressure and mobile phase compositional gradients. In our recent work, we saw that this mode is well suited for large-molecule separations (11). In this combined gradient mode, a positive pressure gradient has been applied, and it is expected to yield a stretched elution window, and thus improved selectivity. However, peaks might suffer from extra band broadening. Meanwhile, a negative pressure gradient is expected to produce a compressed elution window (lower selectivity), but it may result in peak sharpening (11). Figure 3 shows the observed chromatograms when running linear pressure gradient separations. Indeed, the positive pressure gradient resulted in improved selectivity and somewhat broader peaks compared to constant pressure experiments. On the other hand, the negative pressure gradient caused the compression of the elution window, but also sharper peaks, as expected. The peak broadening and compression effects were particularly prominent for the first eluting peak. Its temporal width (at half height) was 0.087 min when applying the positive pressure gradient, whereas it was only 0.042 min (two-fold thinner) in the case of the negative pressure gradient. Table II summarizes the peak widths data observed for the pressure gradient experiments. By all means, pressure gradient separations resulted in significant changes in selectivities and peak widths compared to experiments performed at constant pressure, highlighting the usefulness of pressure gradient experiments.
Figure 3: Gradient elution separations of oligonucleotides performed in (a) positive linear, and (b) negative linear pressure gradient modes. The orange curves show the experimentally measured pressure gradients. Peaks: 1 = dT40; 2 = dT60; 3 = dT80; and 4 = dT100.
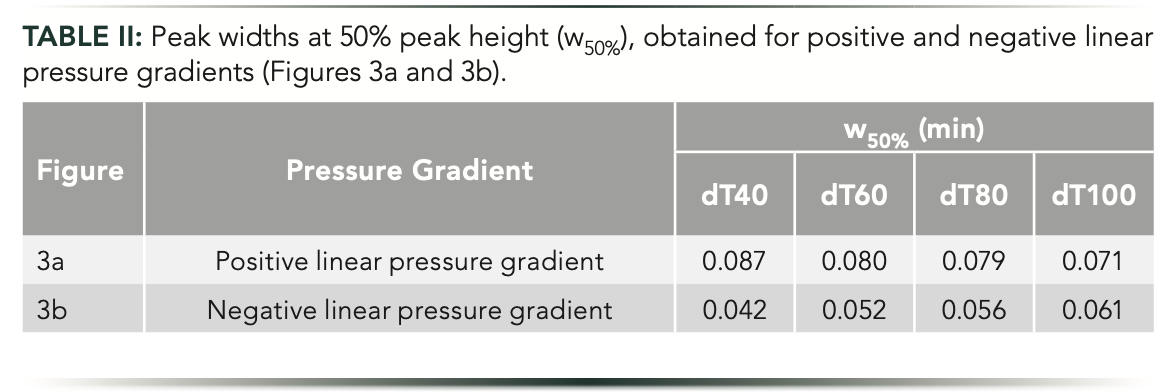
All the pressure-controlled experiments performed in this work suggest that the retention of a given compound can be significantly increased or decreased by changing the operating pressure. This observation motivated us to try a segmented pressure program (stepwise pressure gradient) to selectively shift the retention of a peak or of a group of peaks. The idea is similar to the recently introduced multi-isocratic elution mode of large solutes (14), but using pressure as a controlling source of solute retention instead of mobile phase strength. Figure 4 illustrates this approach applying stepwise pressure programs. At constant pressure, this homopolymer ladder of oligonucleotides shows a typical elution profile for a homolog series (Figure 4a). It is known that when separating homologous series of compounds while performing linear mobile phase gradients, the selectivity decreases as the homolog number (nh; number of interacting functional groups) increases. For homologs with low numbers of functional groups (early eluting peaks), one additional functional group increases drastically the strength of the interaction between the solute and the stationary phase. For homologs possessing high number of functional groups (late eluting peaks), one additional functional group will not increase the strength of their interaction significantly. Therefore, the unequal peak distribution that is often observed in the chromatograms is because of the gradually decreasing molecular differences among the species possessing a higher position in the series (15–17).
Figure 4: Gradient elution separations of oligonucleotides performed with a 450-bar pressure step (F2: 0–0.4 mL/min in 1 min, curve 10) after the elutions of (b) peak 1, (c) peak 2, and (d) peak 3, compared to (a) constant pressure. The orange curves show the experimentally measured pressure gradients. Peaks: 1 = dT40; 2 = dT60; 3 = dT80; and 4 = dT100.
When eluting the first peak at relatively low pressure and then setting a high-pressure step (doubling the inlet pressure), the retention of the late eluting species will be shifted, resulting in significantly increased selectivity between peaks 1 and 2, whereas selectivity between the following eluting peaks will be nearly unchanged (Figure 4b). The same approach can be applied to enhance the selectivity between peaks 2 and 3. The pressure remains unchanged until the elution of the second peak. Then, by adding a pressure step, the retention of the last two eluting species can be selectively shifted (Figure 4c). Finally, the same improvement can be done for the most critical peak pair (peaks 3 and 4). Whatever the mobile phase compositional gradient program, it is hard to obtain a baseline separation for these two peaks. However, with the controlled pressure step program, when setting the ramp right after the elution of the third peak, the elution distance between peaks 3 and 4 can be drastically increased. Without pressure control, Rs ~1.5 could be attained, but with the PE-LC setup, it was possible to reach Rs of 11.8 (Figure 4d). Please note that we are limited at 1000 bar with this PE-LC setup, but if it was possible to set a higher pressure, even greater selectivity could have been achieved.
Evaluating the Possibilities of PE-LC for Intact mRNA Separations in AEX
In most cases, oligonucleotide purification has been performed with either AEX or IP-RP chromatography (18). Indeed, AEX has been regularly used to purify mRNA (19). However, it is interesting to note that only a few analytical separations have been published to date. It is our belief that if the available methods were to be improved (for example, by using low adsorption column hardware and chromatographic systems, and applying new elution modes such as “ion-pairing AEX” [20]), there might be potential for AEX to become a common analytical approach for intact RNA analysis. AEX can be applied with less denaturing conditions than IP-RP; thus, it can preserve the mRNA structure and be potentially used to analyze their native conformational heterogeneity (21).
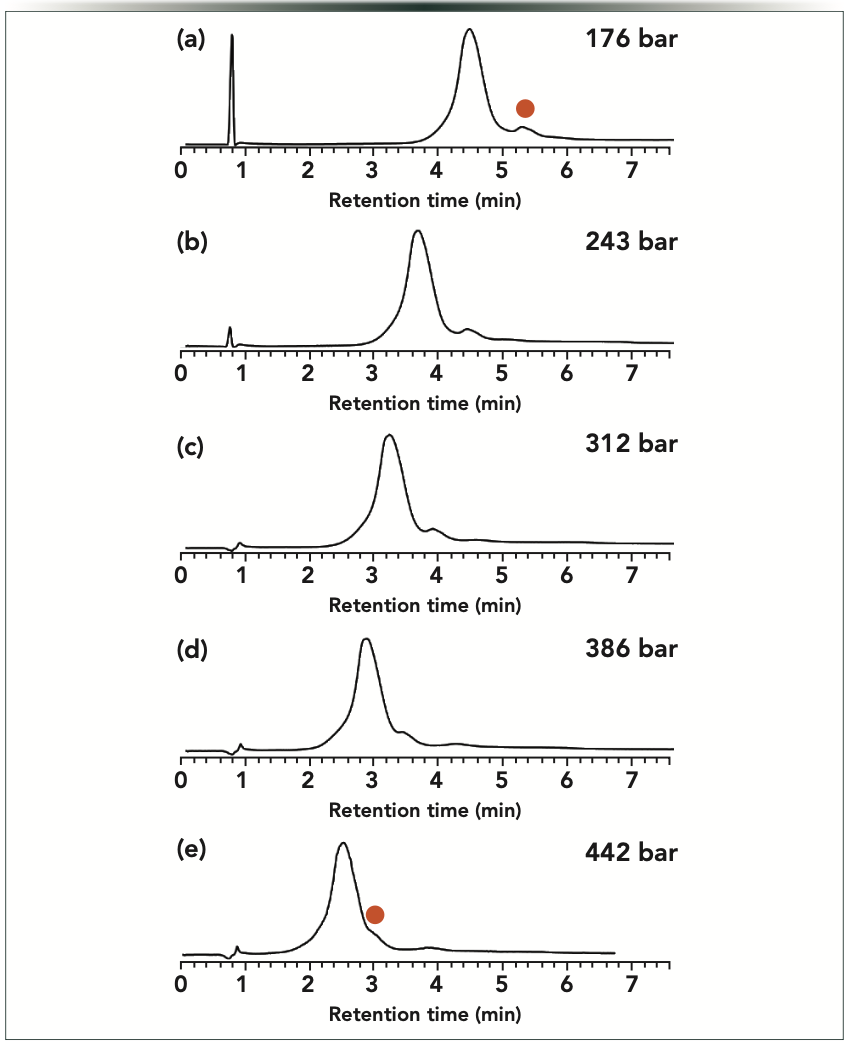
The possibilities offered by PE-LC and direct pressure effects were tested for intact mRNA analysis in AEX. Figure 5 shows the corresponding chromatograms observed for EPO mRNA at various applied pressures. Like for some proteins in IEX, the retention of the intact mRNA decreases with the increase of operating pressure. A major and a minor peak (related to the native heterogeneity of this mRNA sample) can be separated with this AEX method. Interestingly, the selectivity between these two species changes with pressure—the lower the pressure, the higher the selectivity. This observation suggests that even lower pressure (p < 176 bar) would result in an improved selectivity. By decreasing the length of the column (for example, by using the recently introduced ultra-short column formats [22]), it would probably yield an improved separation.
Figure 5: Strong anion exchange gradient elution separations of intact EPO mRNA performed at constant, but different pressures. Experiments were performed at p (bar) = (a) 176, (b) 243, (c) 312, (d) 386, and (e) 442 bar. Orange dot denotes peak for which selectivity changed significantly with pressure.
Studying the Possibilities of PE-LC for the Separation of DNA Ladder Species in AEX
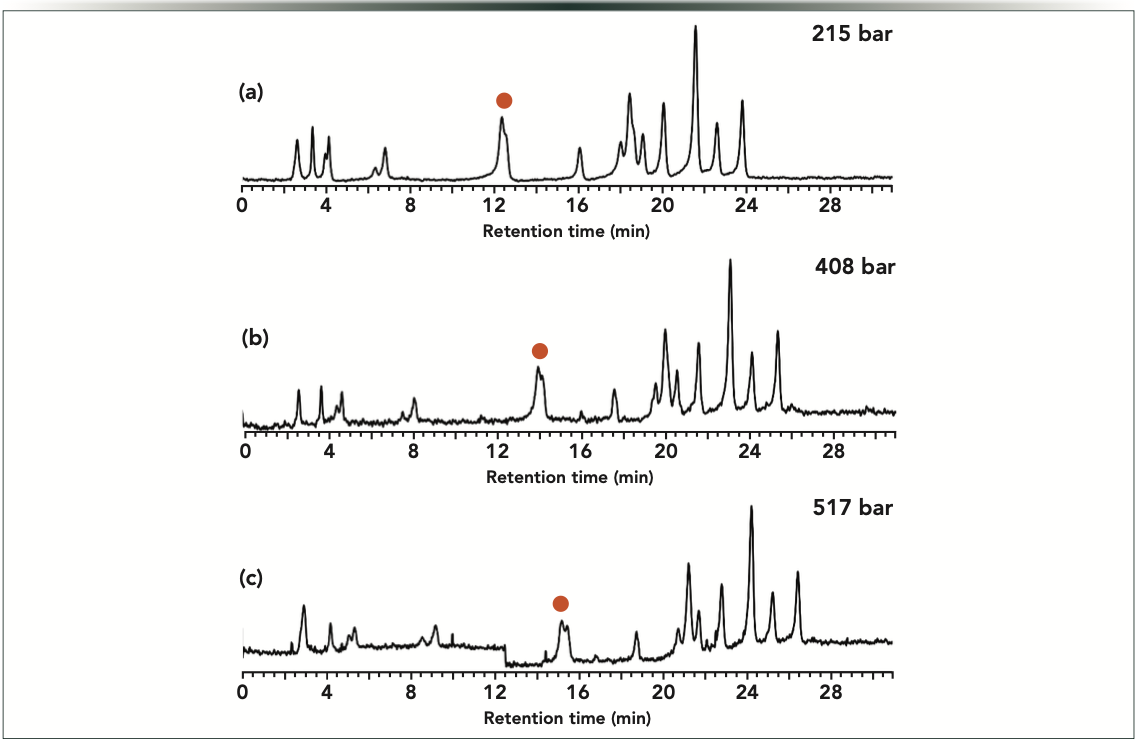
The PE-LC setup was also tested for the separation of DNA species. For such a sample, a slight retention increase was observed at increased pressures. The overall elution profile did not change significantly, but for some of the critical peak pairs, improved selectivity could be attained at the high column head pressure compared to low pressure experiments (Figure 6). In this particular example, only limited pressure effects were seen, but it may happen that larger selectivity effects would be observed in other AEX modes (for example, ion-pairing AEX) or on other stationary phases (for example, weak exchanger compared to strong exchanger).
Figure 6: Strong anion exchange gradient elution separations of 100 bp DNA ladder (NEB, N3231S) performed at constant, but different pressures. Experiments were performed at p (bar) = (a) 215, (b) 408, and (c) 517 bar. The baseline drift was corrected with a linear function. Orange dot denotes peaks for which selectivity improvement was observed with increasing pressure.
Conclusions
Several examples are shown in this study to illustrate the potential of pressure-controlled separations for large molecules (oligonucleotides, mRNAs, DNA species). In most cases, changing the operating pressure or running a pressure gradient (linear gradient or step-wise gradient) result in significant selectivity improvements. However, pressure effects seem to be sample and elution mode dependent, and as such, they are expected to be the object of further investigations.
We believe that this novel PE-LC setup opens new possibilities in LC, especially for moderate to large size analytes. It is worth noting that with the PE-LC approach, unique selectivity can be obtained by intentionally changing the operating pressure (for example, for oligonucleotide separations in IP-RP). Pressure as a LC method variable will increase the degrees of freedom for method development strategies.
Credit Authorship Contribution Statement
Honorine Lardeux helped with the investigation and the experiments for this article. Davy Guillarme reviewed and edited this article. Mateusz Imiołek helped with investigation, experiments, and review of the article. Matthew Lauber helped with the review and editing of the article. Szabolcs Fekete contributed the methodology, investigation, and conceptualization of this study, and he wrote the original article draft.
Acknowledgment
The authors would like to thank Mike Fogwill for his help and support to realize pressure-controlled LC experiments.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Column and LC instruments used in this research were paid for and made available by Waters Corporation. Acquity, UHPLC, and BEH are trademarks of Waters Technologies Corporation.
References
(1) Liu, X.; Szabelski, P.; Kaczmarski, K.; and Zhou, D.; Guiochon, G. Influence of Pressure on the Chromatographic Behavior of Insulin Variants Under Nonlinear Conditions. J. Chromatogr. A. 2003, 988, 205−218. DOI: 10.1016/S0021-9673(03)00002-5
(2) Felinger, A.; Boros, B.; Ohmacht, R. Effect of Pressure on Retention Factors in HPLC Using a Non-Porous Stationary Phase. Chromatographia 2002, 56, S61−S64. DOI: 10.1007/BF02494114
(3) Martin, M.; Guiochon, G. Effects of High Pressure in Liquid Chromatography. J. Chromatogr. A. 2005, 1090, 16−38. DOI: 10.1016/j.chroma.2005.06.005
(4) Fallas, M.M.; Neue, U.D.; Hadley, M.R.; McCalley, D.V.J. Investigation of the Effect of Pressure on Retention of Small Molecules Using Reversed-Phase Ultra-High-Pressure Liquid Chromatography. J. Chromatogr. A 2008, 1209, 195−205. DOI: 10.1016/j.chroma.2008.09.021
(5) Fekete, S.; Veuthey, J.L.; McCalley, D.V.J.; Guillarme, D. The Effect of Pressure and Mobile Phase Velocity on the Retention Properties of Small Analytes and Large Biomolecules in Ultra-High Pressure Liquid Chromatography. J. Chromatogr. A. 2012, 1270, 127−138. DOI:10.1016/j.chroma.2012.10.056
(6) Makarov, A.; LoBrutto, R.; Karpinski, P.; Kazakevich, Y.; Christodoulatos, C.; Ganguly, A.K. Investigation of the Effect of Pressure and Liophilic Mobile Phase Additives on Retention of Small Molecules and Proteins Using Reversed-Phase Ultrahigh Pressure Liquid Chromatography. J. Liq. Chrom. Rel. Technol. 2012, 35, 407−427. DOI:10.1080/10826076.2011.601494
(7) Fekete, S.; Horváth, K.; Guillarme, D. Influence of Pressure and Temperature on Molar Volume and Retention Properties of Peptides in Ultra-High Pressure Liquid Chromatography. J. Chromatogr. A. 2013, 1311, 65−71. DOI: 10.1016/j.chroma.2013.08.045
(8) Fekete, S.; Guillarme, D. Estimation of Pressure-, Temperature- and Frictional Heating-Related Effects on Proteins' Retention Under Ultra-High-Pressure Liquid Chromatographic Conditions. J. Chromatogr. A. 2015, 1393, 73−80. DOI:10.1016/j.chroma.2015.03.023
(9) Liu, X.; Zhou, D.; Szabelski, P.; Guiochon, G. Influence of Pressure on the Retention and Separation of Insulin Variants Under Linear Conditions. Anal. Chem. 2003, 75, 3999−4009. DOI: 10.1021/ac0205964
(10) Martin, M.; Guiochon, G. Effects of High Pressure in Liquid Chromatography. J. Chromatogr. A. 2005, 1090, 16−38. DOI: 10.1016/j.chroma.2005.06.005
(11) Fekete, S.; Fogwill, M.; Lauber, M. Pressure-Enhanced Liquid Chromatography, a Proof of Concept: Tuning Selectivity with Pressure Changes and Gradients. Anal. Chem. 2002, 94, 7877–7884. DOI: 10.1021/acs.analchem.2c00464
(12) Kristl, A.; Caf, M; Pompe, M.; Podgornik, A. Complex Protein Retention Shifts with a Pressure Increase: An Indication of a Standard Partial Molar Volume Increase during Adsorption? Anal. Chem. 2022, 94, 13350–13358. DOI: 10.1021/acs.analchem.2c01809
(13) Fekete, S.; Veuthey, J.L.; Beck, A.; Guillarme, D. J. Hydrophobic Interaction Chromatography for the Characterization of Monoclonal Antibodies and Related Products. Pharm. Biomed. Anal. 2016, 130, 3–18. DOI: 10.1016/j.jpba.2016.04.004
(14) Fekete, S.; Beck, A.; Veuthey, J.L.; Guillarme, D. Proof of Concept to Achieve Infinite Selectivity for the Chromatographic Separation of Therapeutic Proteins. Anal. Chem. 2019, 91, 12954−12961 (2019). DOI: 10.1021/acs.analchem.9b03005
(15) Jandera, P. Simultaneous Optimisation of Gradient Time, Gradient Shape and Initial Composition of the Mobile Phase in the High-Performance Liquid Chromatography of Homologous and Oligomeric Series. J. Chromatogr. A 1999, 845, 133–144. DOI: 10.1016/S0021-9673(99)00331-3
(16) Joshi, V.S.; Kumar, V.; Rathore, A.S. Role of Organic Modifier and Gradient Shape in RP-HPLC Separation: Analysis of GCSF Variants. J. Chromatogr. Sci. 2015, 53, 417–423. DOI: 10.1093/chromsci/bmu222
(17) Bobály, B.; Randazzo, G.M.; Rudaz, S.; Guillarme, D.; Fekete, S. Optimization of Non-Linear Gradient in Hydrophobic Interaction Chromatography for the Analytical Characterization of Antibody-Drug Conjugates. J. Chromatogr. A. 2017, 1481, 82–91. DOI: 10.1016/j.chroma.2016.12.047
(18) Cook, K.; Thayer, J.R. Advantages of Ion-Exchange Chromatography for Oligonucleotide Analysis. Bioanalysis 2011, 3, 1109–1120. DOI: 10.4155/bio.11.66
(19) Gagnon, P. Purification of Nucleic Acids: A Handbook for Purification of Plasmid DNA and mRNA for Gene Therapy and Vaccines; BIA Separations, 2020.
(20) Fekete, S.; Yang, H.; Wyndham, K.; Lauber, M. Salt Gradient and Ion-Pair Mediated Anion Exchange of Intact Messenger Ribonucleic Acids. J. Chromatogr. Open 2022, 2, 100031. DOI: 10.1016/j.jcoa.2022.100031
(21) Fekete, S.; Doneanu, C.; Addepalli, B.; Gaye, M.; Nguyen, J; Alden, B.; Birdsall, R.; Han, D.; Isaac, G.; Lauber, M. Challenges and Emerging Trends in Liquid Chromatography-Based Analyses of mRNA Pharmaceuticals. J. Pharm. Biomed. Anal. 2023, 224, 115174. DOI: 10.1016/j.jpba.2022.115174
(22) Navarro-Huerta, J. A.; Murisier, A.; Nguyen, J. M.; Lauber, M.A.; Beck, A.; Guillarme, D.; Fekete, S. Ultra-Short Ion-Exchange Columns for Fast Charge Variants Analysis of Therapeutic Proteins. J. Chromatogr. A 2021, 1657, 462568. DOI: 10.1016/j.chroma.2021.462568
About the Authors
Honorine Lardeux and Davy Guillarme are with the School of Pharmaceutical Sciences Institute of Pharmaceutical Sciences of Western Switzerland, at the University of Geneva, in Geneva, Switzerland, and the University of Geneva, Geneva, Switzerland. Mateusz Imiołek and Szabolcs Fekete are with Waters Corporation, in Geneva, Switzerland. Matthew A. Lauber is with Waters Corporation, in Milford, Massachusetts. Direct correspondence to: Szabolcs_Fekete@waters.com

Polysorbate Quantification and Degradation Analysis via LC and Charged Aerosol Detection
April 9th 2025Scientists from ThermoFisher Scientific published a review article in the Journal of Chromatography A that provided an overview of HPLC analysis using charged aerosol detection can help with polysorbate quantification.
Removing Double-Stranded RNA Impurities Using Chromatography
April 8th 2025Researchers from Agency for Science, Technology and Research in Singapore recently published a review article exploring how chromatography can be used to remove double-stranded RNA impurities during mRNA therapeutics production.









