Novel Automated Method for Sample Preparation for Peptide Mapping
Peptide mapping is considered a critical quality attribute (CQA) for the identification of a protein’s primary structure and confirmation of any peptide modifications. Peptide mapping can be streamlined with a comprehensive kit and an autolysis-resistant trypsin for high-efficiency digestions. Automation enhances laboratory productivity and reduces errors in peptide mapping, making it advantageous to implement in a biopharmaceutical laboratory.
If you have been tasked with developing a peptide mapping method to further characterize your protein, what is peptide mapping and where do you begin? Peptide mapping is the analysis of chemical or enzymatic digests of a protein to focus on specific peptide regions and modifications (1). It is an important assay in biopharmaceutical characterization because it helps identify the primary protein structure to confirm process consistency and genetic stability (1). Many scientists think of a peptide map as a “fingerprint” of a protein that provides a comprehensive understanding of the protein being analyzed (1).
The entire peptide mapping workflow can be broken down into three major steps: sample preparation, separation by reversed-phase liquid chromatography, and, lastly, data processing. Each step presents its own set of challenges, with sample preparation being highly laborious, complex, and prone to variability.
Traditionally, there are five key steps involved in preparing samples for peptide mapping: denaturation, reduction, alkylation, desalting, and digestion. It is easy to imagine the many ways that error and variability could be introduced in each step when developing a robust sample preparation method. Among the considerations that must be evaluated are reagent reliability, desalting inconsistencies, digestion completion, and enzyme autolysis. Beyond these, it is important to consider the time required to perform the peptide mapping sample preparation workflow. Depending on the length of digestion, the entire sample preparation method can be more than 8 h. These long digestion times often also introduce artificial peptide modifications, such as asparagine deamidation and methionine oxidation (2).
This process can be laborious and overwhelming—which is why it is no surprise that many are looking to automated processes to streamline this work. The use of an automated liquid handler for increasing throughput and alleviating the time spent on routine pipetting is of great interest to scientists, especially for sample preparation for complex assays such as peptide mapping. Historically, this type of automation has proven difficult, requiring large investments of both time and money, as well as introducing new skill sets such as expertise in Python scripting. However, recent advances in the development of new, automated solutions with intuitive software can enable total, “hands-free” solid-phase extraction.
Protein denaturation, reduction, and alkylation are performed to provide the proteolytic enzyme access to digestion sites for a more complete digestion (3). The most common denaturant used in peptide mapping is guanidine hydrochloride. If trypsin is used to cleave peptide bonds, guanidine can significantly interfere with its activity (3). Therefore, a desalting step is needed to effectively remove the guanidine prior to digestion (3). Size-exclusion chromatography (SEC), also known as gel filtration, is a common desalting mechanism where analytes are separated based on their size in solution. Larger molecular weight analytes pass through the chromatographic bed and are eluted first while alkylating reagents are trapped in the pores of the desalting device, which allows the larger denatured, reduced, and alkylated protein to flow through first, as depicted in Figure 1.
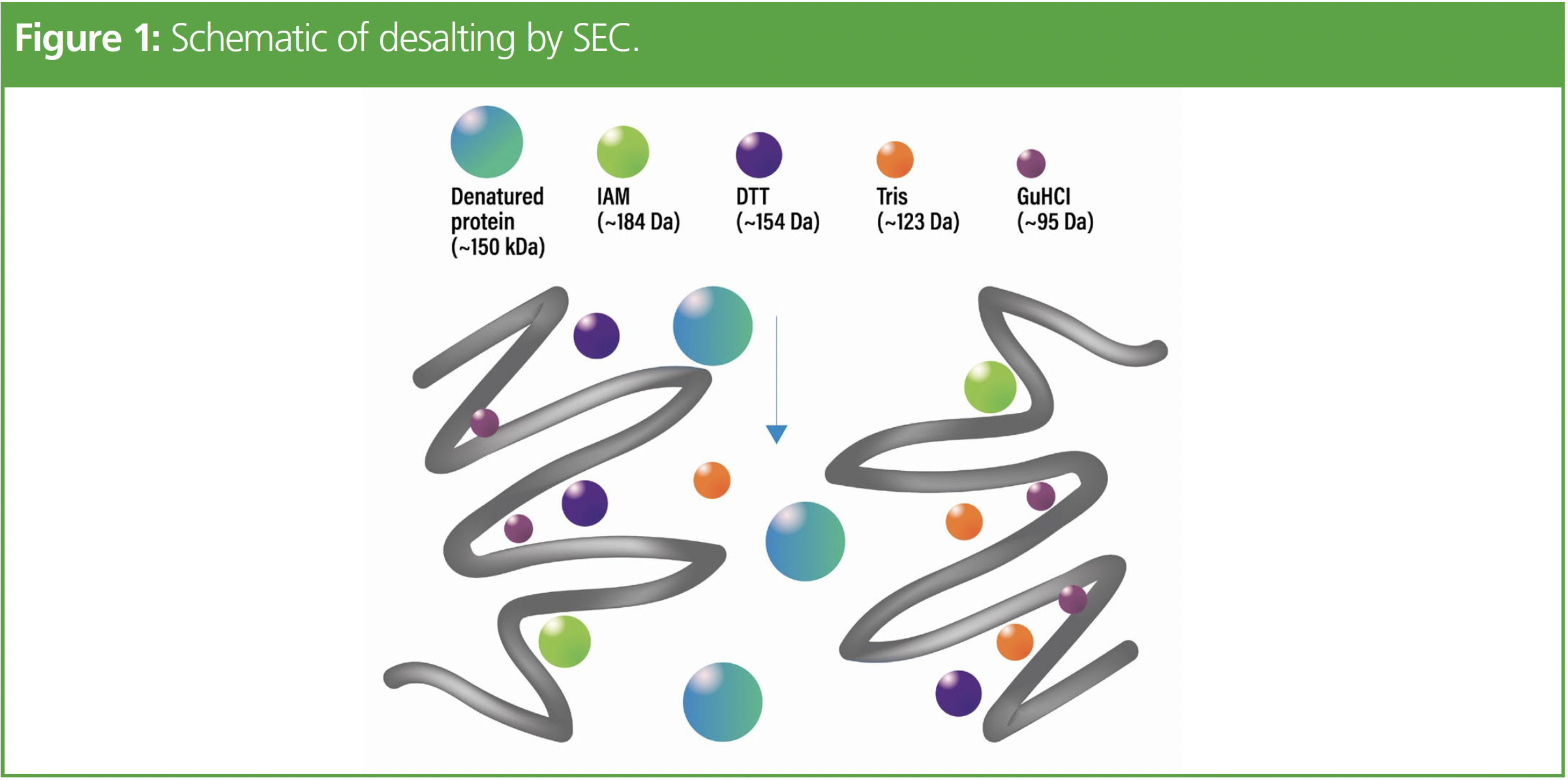
Desalting devices can be in a 96-well plate, microcentrifuge tube, or cartridge format. While easy to automate, plate-based desalting devices suffer from limited loading capacity. On the contrary, the use of microcentrifuge tube-based devices can be more cumbersome and difficult to automate as a centrifuge is often required to process the samples. Cartridge-based desalting devices rely on gravity for elution, but use of these devices is difficult to automate and integrate with high-throughput workflows. Recent advances in the design and development of desalt cartridges allow for use of a multichannel pipette in a 96-well stand for ease-of-use and amenability to automation platforms (4).
Last but not least is the daunting task of developing a robust enzymatic digestion procedure. Trypsin, the most common enzyme for peptide mapping, cleaves proteins on the C-terminal side of lysine and arginine (5). The elevated temperature and alkaline pH used in tryptic digestions can induce artificial peptide modifications, complicating data analysis (5). Tryptic digestions are further complicated by missed cleavages (under digestion), non-specific cleavages (over-digestion), and autolysis (when an enzyme starts to digest itself).
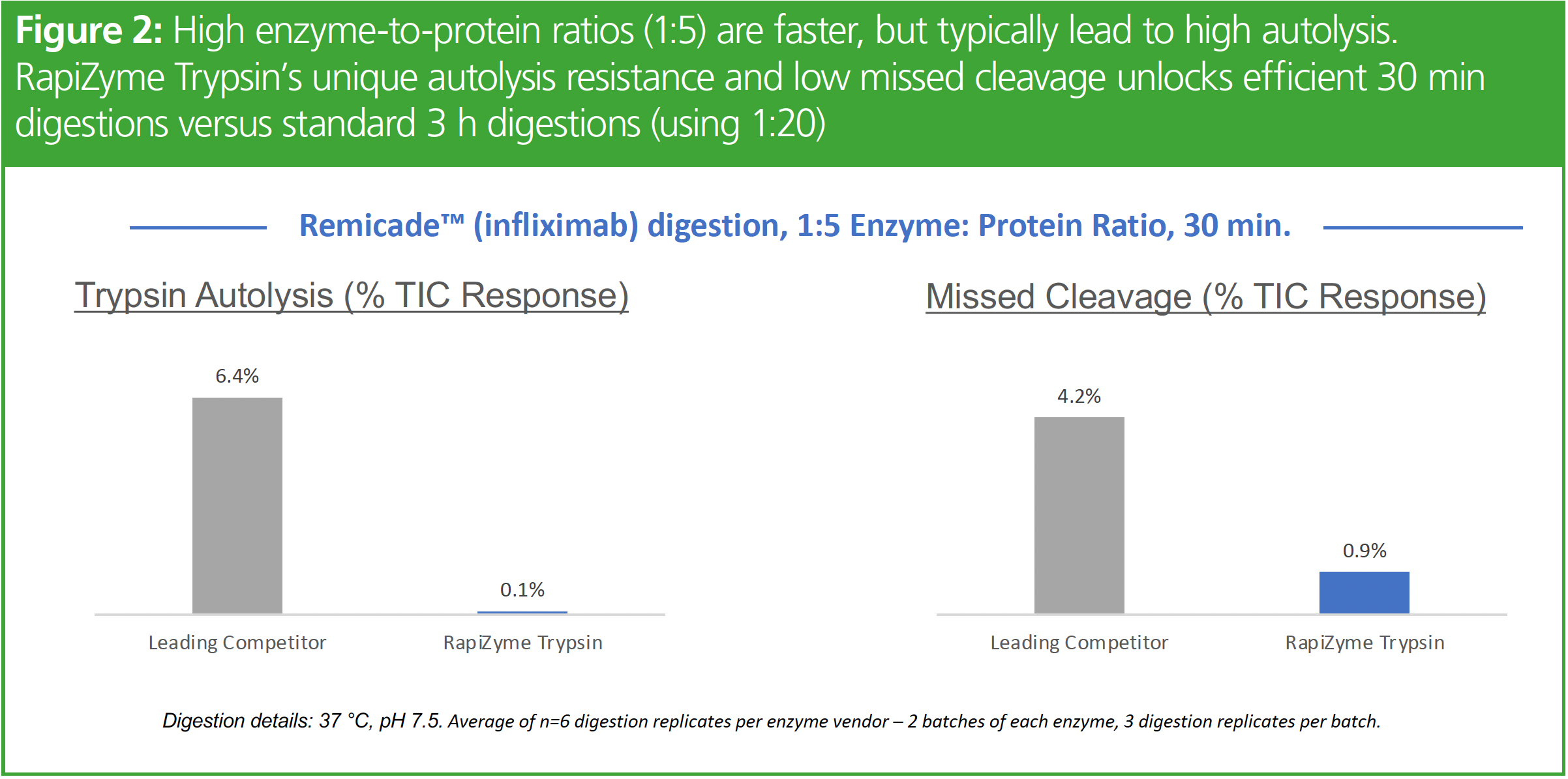
A novel, autolysis-resistant, homogenously methylated recombinant porcine trypsin was recently introduced to help overcome the challenges in tryptic digestion (6). This trypsin enables fast 30-min digestions using 1:5 enzyme:protein ratio with minimal autolysis and low missed cleavages as shown in Figure 2.
Method
NIST Monoclonal Antibody (NISTmAb) Reference Material 8671 was digested using a comprehensive peptide mapping kit, PeptideWorks Tryptic Protein Digestion Kit, which includes RapiZyme Trypsin. NISTmAb reference tryptic digest samples were prepared both manually and with automation on the Andrew+ Pipetting Robot with Extraction+ Connected Device. NISTmAb samples (10 mg/mL) were denatured and reduced in a solution containing 5 M guanidine hydrochloride (GuHCl) and 5 mM dithiothreitol (DTT) for 30 min at room temperature. Iodoacetamide (IAM) was then added to a final concentration of 10 mM and the samples were incubated for 30 min at room temperature in the dark. Samples were desalted with Sep-Pak SEC Desalting Cartridges and buffer exchanged with digestion buffer (10 mM CaCl2 and 100 mM Tris HCl, pH 7.5). The concentration of desalted samples was measured with a UV plate reader and normalized to 0.1 mg/ mL using the digestion buffer as a diluent. RapiZyme Trypsin was added to each sample at a 1:5 enzyme:protein ratio and digestion proceeded for 30 min at 37 °C. Finally, the reaction was quenched with 1% formic acid to a final concentration of 0.1% and injected onto the LC–MS system.
Results
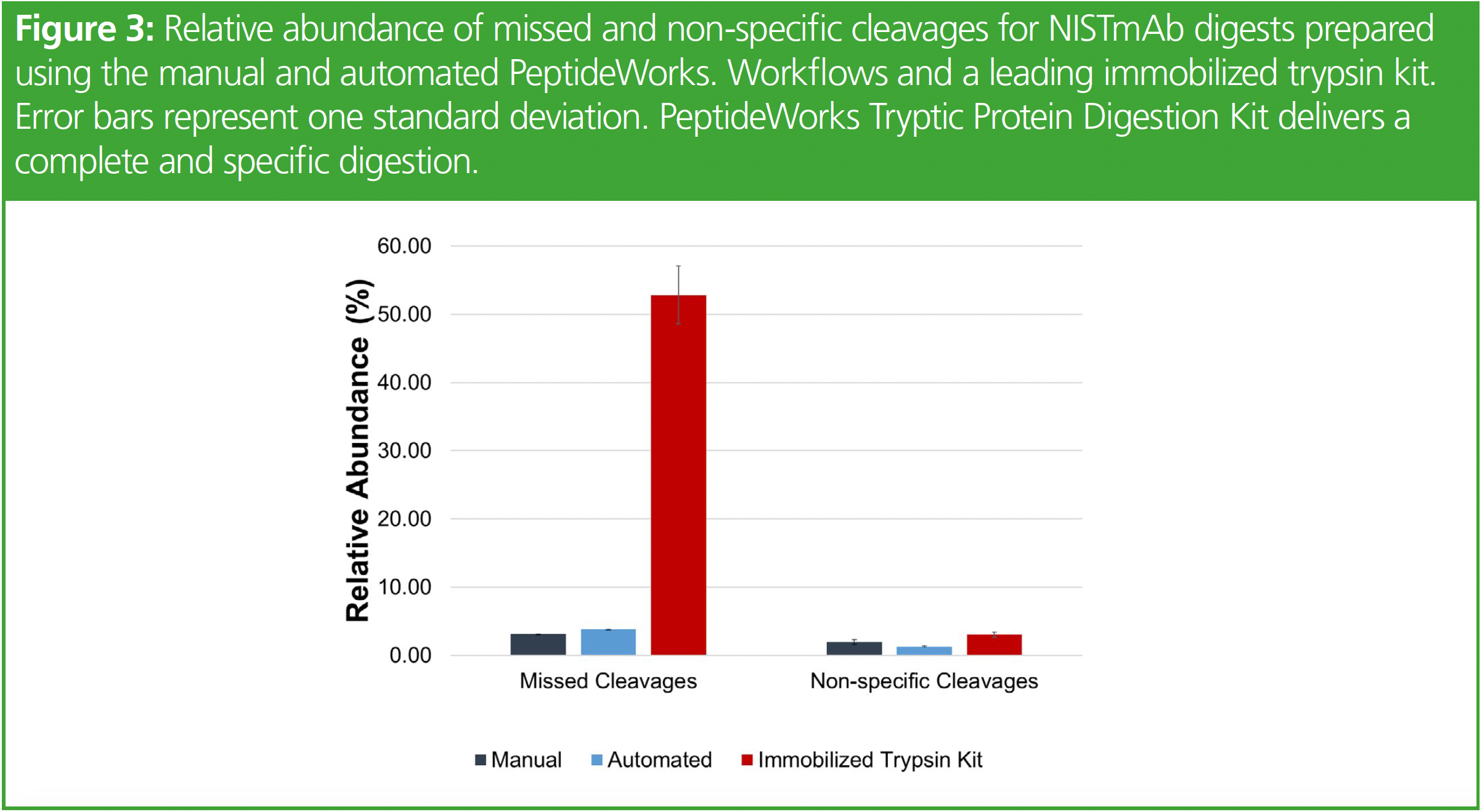
Figure 3 displays the relative missed and non-specific cleavage results for NISTmAb digests prepared using manual and automated. The automated workflow used delivers NISTmAb digests with less than 5% missed and non-specific cleavages, indicating high digestion efficiency without over-digestion of the protein. Missed and non-specific cleavage results for NISTmAb digests prepared using an immobilized trypsin digest kit are based on published results and shown in red in Figure 3. The digestion kit used yields a 93% reduction in missed cleavages and 55% reduction in non-specific cleavages compared to the immobilized trypsin digest kit (7).
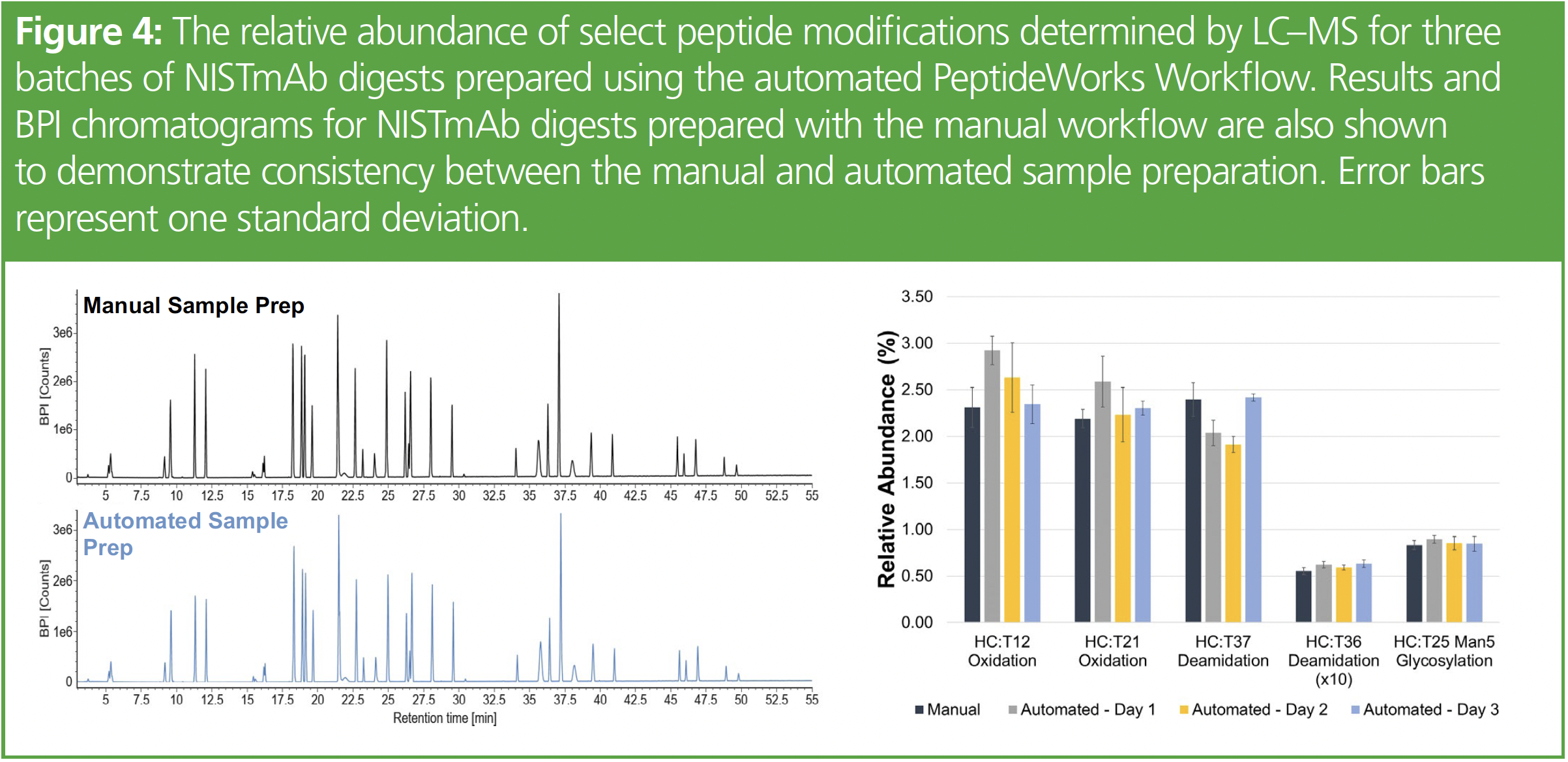
BPI chromatogram overlays of NISTmAb digests prepared using the manual and automated workflows (left) and the relative abundance of select peptide modifications for three batches (right) on the pipetting robot are shown in Figure 4. These results show that a comprehensive peptide mapping kit provided fast tryptic digests without sacrificing digestion completion or inducing high levels of method-induced deamidation or oxidation. The optimized automation protocol generates reproducible results over time and provides an increase in productivity with simple, robust automation alleviating potential pipetting errors (5).
Conclusion
Developing a robust sample preparation method for peptide mapping can be laborious and filled with many variables. Faster digestions often yield incomplete digests with additional autolysis species making data analysis more complex. Peptide mapping can be streamlined using a comprehensive kit with lot tracible reagents and a novel autolysis-resistant high purity trypsin allowing for high-efficiency 30 min digestions. The use of automation can further enhance laboratory productivity and reduce errors, especially in a labor-intensive method, such as peptide mapping.
Acknowledgment
Special thanks to Bill Warren, Caitlin Hanna, Meagan Callis and Stephan Koza for your contributions.
References
(1) USP. Biotechnology-Derived Articles–Peptide Mapping <1055>; USP32–NF27 Page 496, Interim Revision Announcement: USP32–NF27 No. 1 Page 41, Pharmacopeial Forum: Volume No. 32(2) Page 571; Rockville, MD, 2016. https://www.usp.org/sites/default/files/usp/document/harmonization/biotechnology/b05_pf_ira_35_1_2009.pdf (accessed 2023-12-11).
(2) Jiang, P.; Li, F.; Ding, J. Development of an Efficient LC–MS Peptide Mapping Method Using Accelerated Sample Preparation for Monoclonal Antibodies. J. Chromatogr. B 2020, 1137, 121895. DOI: 10.1016/j.jchromb.2019.121895
(3) Mouchahoir, T.; Schiel, J. E. Development of an LC–MS/MS Peptide Mapping Protocol for the NISTmAb. Anal. Bioanal. Chem. 2018, 410, 2111–2126. DOI: 10.1007/s00216-018-0848-6
(4) Sep-Pak SEC Desalting Cartridges, 1cc 5K MWCO Care and Use Manual. Waters User Manual 720007983EN; Waters Corporation: Milford, MA, 2023. https://www.waters.com/webassets/cms/support/docs/720007983en.pdf (accessed 2024-02-21).
(5) Ren, D.; Pipes, G. D.; Liu, D.; et al. An Improved Trypsin Digestion Method Minimizes Digestion-Induced Modifications on Proteins. Anal. Biochem. 2009, 392 (1), 12–21. DOI: 10.1016/j.ab.2009.05.018
(6) Ippoliti, S.; Zampa, N.; Yu, Y. Q.; Lauber, M. A. Versatile and Rapid Digestion Protocols for Biopharmaceutical Characterization Using RapiZyme Trypsin; Waters Application Note 720007840; Waters Corporation: Milford, MA, 2023. https://www.waters.com/nextgen/us/en/library/application-notes/2023/versatile-and-rapid-digestion-protocols-for-biopharmaceutical-characterization-using-rapizyme-trypsin.html (accessed 2024-02-21).
(7) Hanna, C. M.; Danaceau, J. P.; Koza, S. M.; Shiner, S., Trudeau, M. Quick and Robust Sample Preparation for Tryptic Peptide Mapping with the PeptideWorks Kit using Simple, Automatable Workflows; Waters Application Note 720008019; Waters Corporation: Milford, MA, 2023. https://www.waters.com/nextgen/us/en/library/application-notes/2023/quick-robust-sample-preparation-fortryptic-peptide-mapping-with-the-peptideworks-kit-using-simple-automatable-workflows.html (accessed 2024-02-20).
Leslie Napoletano is a Principal Product Marketing Manager within Waters Consumables & Lab Automation Group. She has been with Waters since 2012 providing support and education to scientists in New Jersey with Waters chemistry consumables. More recently in 2021, Leslie has moved into the Product Marketing group focusing on bringing on new consumable products for protein characterization. Prior to joining Waters, Leslie worked as a scientist in protein characterization in the pre-clinical development of monoclonal antibodies at Merck and prior to that at Teva as a R&D chemist. Email: Leslie_Napoletano@waters.com

Study Explores Thin-Film Extraction of Biogenic Amines via HPLC-MS/MS
March 27th 2025Scientists from Tabriz University and the University of Tabriz explored cellulose acetate-UiO-66-COOH as an affordable coating sorbent for thin film extraction of biogenic amines from cheese and alcohol-free beverages using HPLC-MS/MS.
Multi-Step Preparative LC–MS Workflow for Peptide Purification
March 21st 2025This article introduces a multi-step preparative purification workflow for synthetic peptides using liquid chromatography–mass spectrometry (LC–MS). The process involves optimizing separation conditions, scaling-up, fractionating, and confirming purity and recovery, using a single LC–MS system. High purity and recovery rates for synthetic peptides such as parathormone (PTH) are achieved. The method allows efficient purification and accurate confirmation of peptide synthesis and is suitable for handling complex preparative purification tasks.