Multidimensional Gas Chromatography: Benefits and Considerations for Current and Prospective Users
Two-dimensional gas chromatography (GC×GC) offers improved separation power for complex samples containing hundreds to thousands of analytes. However, several considerations must be made to determine whether multidimensional gas chromatography (MDGC) is the logical instrument choice to answer a particular scientific question, including, but not limited to, whether the analysis is targeted or non-targeted, the number of analytes of interest, and the presence of interferences that are coeluted, as well as any potential regulatory or industrial constraints. Currently, MDGC remains daunting for many users because of data complexity and the limited tools commercially available, which are critical for improving the accessibility of MDGC. Herein, we discuss considerations that may assist analysts, laboratory managers, regulatory agents, instrument and software vendors, and those interested in understanding the applicability of 2D-GC for the scientific question being investigated.
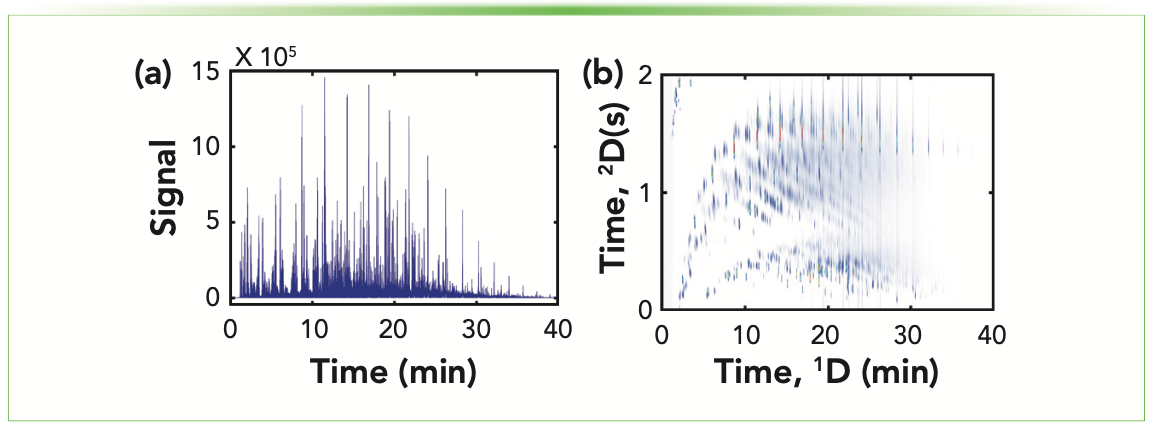
Since the introduction of modern gas chromatography (GC) in the 1950s, GC has been established as the premier technology for the analysis of volatile and semi-volatile organic compounds because of its ease of use, rugged design, and separation power (1). GC is employed across research, regulatory, and industrial applications, requiring the development of more powerful instrumental and data analysis approaches to keep pace with the wealth of complex samples that require analysis. One such instrumental improvement aims to increase peak capacity by employing a second GC separation dimension, referred to as multidimensional gas chromatography (MDGC). The most popular form of MDGC, and the type discussed herein, is comprehensive two-dimensional GC (GC×GC), a technique pioneered approximately 30 years ago by Liu and Phillips (2) that offers an order of magnitude higher peak capacity when method run times are held constant (3–7). For samples such as the diesel fuel as seen in Figure 1, the alkane band dominates the one-dimensional (1D) separation in Figure 1a. The addition of a second column separates alkanes in the 2nd (y) dimension, increasing peak capacity, and allowing improved separation of more trace level analytes, such as aromatics, as demonstrated in Figure 1b.
FIGURE 1: (a) 1D-GC separation of diesel fuel, demonstrating the challenges of complex sam- ples and ability to resolve coeluting analytes. (b) GC×GC separation of the same diesel sample. On the second dimension, aromatics elute between 0 and 0.50 s, alkenes and cycloalkanes from 0.5–1.25 s, and the alkane band between ~1.25–2 s.
With the invention of more powerful instrumentation comes the increased responsibility of clearly defining the problem to be addressed, and then carefully selecting the best analytical approach, whether it be GC×GC or 1D-GC with an appropriate detector, be it a mass spectrometer or otherwise. In general, there are several considerations that should be made when performing GC separations, including sample preparation and introduction (8–12), hardware and software selection (5,13,14), and more (15). This article will specifically focus on the chromatographic questions and considerations that need to be made regarding whether to perform one- or two-dimensional GC.
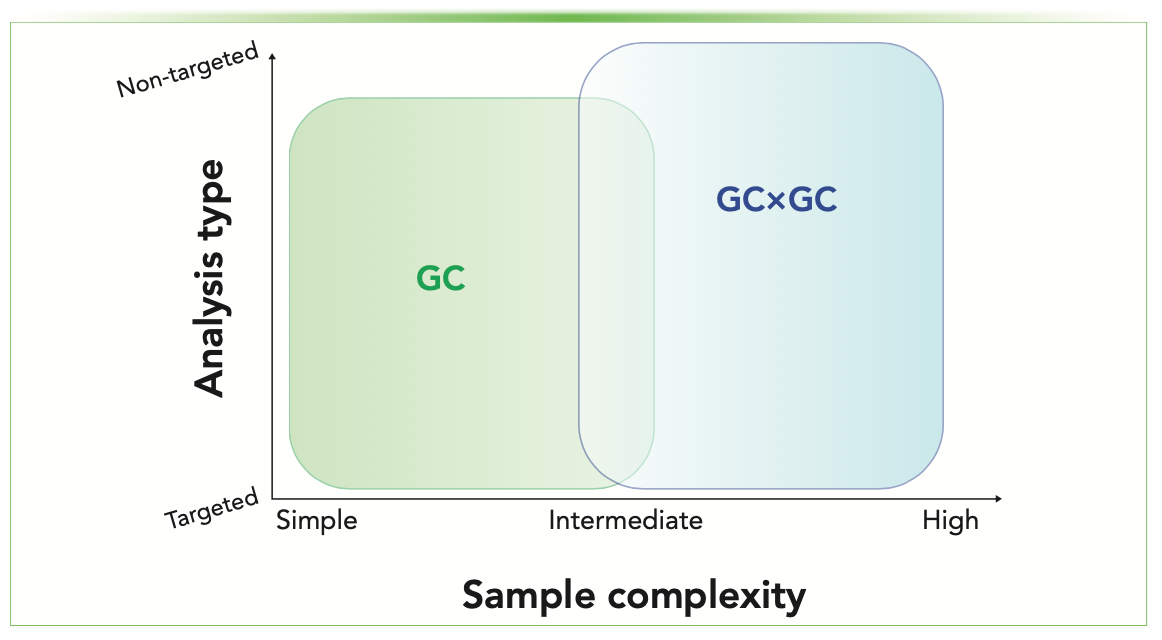
The best technique for any analytical problem depends on multiple factors, such as: 1) whether the study is targeted or non-targeted; 2) if targeted, the number of target analytes to be separated; 3) whether there are interferents in the sample from the extraction process, stationary phase background, or matrix that are co-eluted with the analytes of interest; 4) the advantages or limitations of available detectors; and 5) resources, such as analyst training and availability, time, money, and other regulatory or industrial constraints. For example, as can be observed in Figure 2, quantification of preservatives in cosmetic products would likely fall in the bottom left corner of the diagram because of the relatively low sample complexity and targeted quantification (16). In contrast, a differential comparison between similar matrix samples would likely benefit from the increased peak capacity of GC×GC (3,17,18).
FIGURE 2: Summary of types of analyses and when GC or GC×GC should be used. For samples of simple complexity, GC is often sufficient. However, as sample complexity increases, GC×GC can provide enhanced separation power for targeted and untargeted analysis.
Comprehensive Multidimensional Gas Chromatography: Strengths and Benefits
In the case of highly complex samples (those that contain more than 150–250 relevant compounds, or have complex matrices not removed via sample preparation), the separation power of traditional 1D-GC decreases because of limited peak capacity (7). As observed in Figure 1b, the addition of a second separation dimension, when shown in a contour plot, enables both x- and y-direction time separations. Consistently co-eluted compounds, especially low-boiling point compounds, can now be separated by the second dimension (generally polarity differences) in addition to the primary separation (generally mobile-to-stationary phase affinity differences). When complementary stationary phases are selected, the second dimension enables a large increase in available space for analytes to occupy. In terms of raw peak capacity, multidimensional chromatography can provide a ~10-fold increase in peak capacity (17).
Complementary stationary phase selection can not only dramatically increase peak capacity of the entire separation, but it also allows chromatographers to independently determine the stationary phase of the primary or secondary column, which is selective for the analytes of interest. For example, a 100% dimethylpolysiloxane primary column, coupled with a secondary column containing 8% phenyl polycarborane-siloxane, will allow for polychlorinated biphenyl (PCB) compounds to be separated according to ortho ring substitution (19). Additionally, GC×GC chromatograms are highly structured according to compound type, as evidenced in Figure 1b of a diesel sample. Alkanes are eluted in a discrete band and are separate from the aromatic region, for example (20–22). Ultimately, one of the advantages of GC×GC is its ability to provide valuable information about probable compound structure and isomer identity, even when an analytical standard is unavailable or library match is poor (19,23–27).
The heart of a GC×GC instrument is the modulator, which, in essence, acts as a second injector onto the shorter, second dimension. Regardless of the type, the modulator traps and compresses analytes prior to the detector, which increases signal to noise (S/N) and can improve limits of detection (LOD) (3–5,28–30). In the case of cryogenic jet-based modulators, peak compression in the modulator can impart a three- to nine-fold increase in analyte sensitivity compared to 1D-GC (17,31). This is of particular interest for environmental analyses, where scientists are asked to detect ever-decreasing levels of analytes (such as perfluoroalkyls and other persistent organic pollutants [POPs]), or when the target presents a health concern, as in the cases of some carcinogens and mutagens (19,23,26,32–34). Furthermore, the oven ramp programs for many chromatographic separations often reach a plateau, causing late-eluting analytes to broaden and lose S/N. The modulator can assist detection by increasing S/N for analytes at the edge of detectability for 1D-GC (35,36).
The evidence is clear that GC×GC is an effective tool for complex analyses, non-targeted analyses, and low detection limits, and could be deployed more universally in laboratories. However, cases exist where, despite its clear advantages, GC×GC is unnecessary for accurate and timely achievement of the analytical goal at hand.
The Flip Side: When 1D-GC is Sufficient or Preferred
The largest subset of analyses that are not suitable for GC×GC consist of small, well-defined problem sets. These include targeted analyses that already have well-established methods, analyses occurring in simple sample matrices that are easily separated from the compounds of interest, or analyses in which a suitably selective detector already exists. For example, quantitative methods confirming ingredient-level concentrations of safe targets, such as preservatives in cosmetic products or caffeine in cocoa, generally have well-established methods using GC or LC coupled with either an univariate or multivariate detector (37,38). Even some cases of increasing complexity can be easily analyzed by chromatography coupled with an appropriate mass spectrometer when a unique m/z or appropriate multiple reaction monitoring (MRM) transition can increase the selectivity and sensitivity of the target over the matrix, such as the analysis of pesticides by GC-triple quadrupole mass spectrometry (TQMS) (37). Detectors that are selective for certain compound classes, such as sulfur chemiluminescence, can provide the selectivity needed for a method to differentiate between the sample and target (39–41). When such techniques are available, and additional information about the sample is not needed, they should be employed over utilizing GC×GC.
There has been increased interest within the GC×GC field regarding the development of GC×GC instruments with other mass spectrometer types (18,42–44). However, a limiting factor that may exclude GC×GC from use with MS is the scan rate required for effective sampling of modulated peaks. These peak widths can be as low as 50 ms (29). To effectively sample such narrow peaks for quantification, a scan rate of at least 100 Hz is generally required. Older detectors and higher resolution instruments do not have this fast, broad-range scan rate (45–47). For those instruments that do have the necessary scan rate, higher resolution data can impart upon GC×GC data a level of complexity beyond which most analysts can manipulate and interpret. Whether it is a hardware or software limitation, detector selection must be suitable for GC×GC analysis—from separation to data interpretation.
Raw GC×GC data can be complex and difficult to interpret, especially in non-targeted work. Consequently, analysts must choose between the following: 1) manual interpretation; 2) vendor software; and 3) in-house data analysis tools. Depending on the complexity and volume of data, manual interpretation may be tedious and impractical for industrial or regulatory laboratories. Although there has been a large amount of growth with vendor data analysis software in recent years, there exists a need to expand the abilities of each software platform and make these tools accessible to a broad audience (48–50). Currently, many GC×GC-focused analysts have turned to the development of in-house chemometric tools to bridge the gap between vendor software (5,13,14,51–54). However, this generally requires the knowledge of at least one scripting language (such as Python or Matlab) and a strong understanding of the statistics-based and chemometric algorithms that aid in the manipulation of GC×GC data and the elucidation of important features within the data. Ultimately, chromatographers now need not only a deep understanding of analytical principles, but they also need a working knowledge of data science.
Beyond the chiefly analytical reasons not to employ GC×GC, there are more practical reasons some avoid the technique, namely, time and resources. GC×GC requires time and expertise to set up, maintain, optimize, and validate GC×GC systems and methods. For urgent industrial problems, there may not be time to employ GC×GC when same-day answers are needed. For non-targeted discovery, GC×GC can be an invaluable tool, but many will move from non-targeted GC×GC to targeted heart-cutting MDGC or 1D as soon as targets are identified for simplicity and allocation of resources (47,55). For example, a GC×GC system can take up valuable resources, whether it be bench space for a dedicated GCxGC system in the laboratory, liquid nitrogen for a cryogenic modulator, or an experienced analyst to maintain the system and analyze the data. Although R&D teams may be able to simply "turn off" the modulator and run a GC×GC system as 1D, such modifications of validated methods may not be permitted in regulatory or industrial environments (56,57).
Conclusion
Ultimately, the principles and considerations when selecting a GC platform can be influenced by a researcher’s own implicit bias related to GC×GC. Experienced members of the GC×GC community have seen firsthand the incredible separation power of these instrument platforms, especially when higher order chemometric analysis is desired. The danger in this mentality is the reality that not all scientific questions require an additional separation dimension. However, although the use of GC×GC can provide valuable information regarding complex samples, it has not gained widespread use thus far. Perhaps a major obstacle for the implementation of GC×GC in many laboratories involves the inherent complexity of the data produced (14). Ultimately, MDGC provides multiple dimensions of data—primary and second dimension separation times, and if a mass spectrometer is used as a detector, mass spectral information. Data analysis may seem daunting, especially in the cases of large data sets, and when coupled with the relative infancy of many chemometric software platforms, may delay the desire to fully integrate GC×GC into regulatory, industrial, and forensic laboratories (18). Undoubtedly, GC×GC would benefit from continued investment in GC×GC hardware and software tools, as well as effective communication, to ultimately bridge the gap between chemistry and data science, and increase accessibility of these powerful separations.
References
(1) M.J.E. Golay, Anal. Chem. 29(6), 928–932 (1957). DOI: https://doi.org/10.1021/ac60126a019.
(2) Z. Liu, J.B. Phillips, J. Chromatogr. Sci. 29(6), 227–231 (1991). https://doi.org/10.1093/chromsci/29.6.227.
(3) L.M. Blumberg, F. David, M.S. Klee, and P. Sandra, J. Chromatogr. A 1188(1), 2–16 (2008). https://doi.org/10.1016/j.chroma.2008.02.044.
(4) J.V. Seeley and S.K. Seeley, Anal. Chem. 85(2), 557–578 (2013). https://doi.org/10.1021/ac303195u.
(5) S.E. Prebihalo, K.L. Berrier, C.E. Freye, H.D. Bahaghighat, N.R. Moore, D.K. Pinkerton, and R.E. Synovec, Anal. Chem. 90(1), 505–532 (2018). https://doi.org/10.1021/acs.analchem.7b04226.
(6) M.S.S. Amaral, Y. Nolvachai, and P.J. Marriott, Anal. Chem. 92(1), 85–104 (2020). https://doi.org/10.1021/acs.analchem.9b05412.
(7) J.B. Phillips and J. Beens, J. Chromatogr. A 856(1), 331–347 (1999). https://doi.org/10.1016/S0021-9673(99)00815-8.
(8) D.W. Potter and J. Pawliszyn, Environ. Sci. Technol. 28(2), 298–305 (1994). https://doi.org/10.1021/es00051a017
(9) S.J. Lehotay, J. AOAC Int. 90(2), 485–520 (2007). https://doi.org/10.1093/jaoac/90.2.485.
(10) N. Reyes-Garcés, E. Gionfriddo, G.A. Gómez-Ríos, Md.N. Alam, E. Boyacı, B. Bojko, V. Singh, J. Grandy, and J. Pawliszyn, Anal. Chem. 90(1), 302–360 (2018). https://doi.org/10.1021/acs.analchem.7b04502.
(11) S.C. Moldoveanu and V. David, “Derivatization Methods in GC and GC/MS,” IntechOpen, 2018. https://doi.org/10.5772/intechopen.81954.
(12) D.R. Knapp, J. Pharm. Sci. 69(5), 619–619 (1980). https://doi.org/10.1002/jps.2600690549.
(13) P.-H. Stefanuto, A. Smolinska, and J.-F. Focant, TrAC Trends Anal. Chem. 139, 116251 (2021). https://doi.org/10.1016/j.trac.2021.116251.
(14) K.L. Berrier, S.E. Prebihalo, R.E. Synovec, Separation Science and Technology 12, 229–268 (2020). https://doi.org/10.1016/B978-0-12-813745-1.00007-6.
(15) J.-M. D. Dimandja, Separation Science and Technology 12, 1–40 (2020). https://doi.org/10.1016/B978-0-12-813745-1.00001-5.
(16) C. Azorín, J.L. Benedé, A. Chisvert, and A. Salvador, A. J. Pharm. Biomed. Anal. 209, 114493 (2022). https://doi.org/10.1016/j.jpba.2021.114493.
(17) M.S. Klee, J. Cochran, M. Merrick, and L.M. Blumberg, J. Chromatogr. A 1383, 151–159 (2015). https://doi.org/10.1016/j.chroma.2015.01.031.
(18) L.M. Dubois, S. Aczon, J.-F. Focant, and K.A. Perrault, Anal. Chem. 92(14), 10091–10098 (2020). https://doi.org/10.1021/acs.analchem.0c01926.
(19) J.-F. Focant, A. Sjödin, W.E. Turner, and D.G. Patterson, Anal. Chem. 76(21), 6313–6320 (2004). https://doi.org/10.1021/ac048959i.
(20) B.A. Parsons, D.K. Pinkerton, B.W. Wright, and R.E. Synovec, J. Chromatogr. A 1440, 179–190 (2016). https://doi.org/10.1016/j.chroma.2016.02.067.
(21) B. Kehimkar, B.A. Parsons, J.C. Hoggard, M.C. Billingsley, T.J. Bruno, and R.E. Synovec, Anal. Bioanal. Chem. 407(1), 321–330 (2015). https://doi.org/10.1007/s00216-014-8233-6.
(22) B.C. Reaser, B.W. Wright, and R.E. Synovec, Anal. Chem. 89(6), 3606– 3612 (2017). https://doi.org/10.1021/acs.analchem.6b04991.
(23) E. Skoczyńska, P. Korytár, and J. de. Boer, Environ. Sci. Technol. 42(17), 6611–6618 (2008). https://doi.org/10.1021/es703229t.
(24) F. Stilo, C. Bicchi, A.M. Jimenez-Carvelo, L. Cuadros-Rodriguez, S.E. Reichenbach, and C. Cordero, Trends Anal. Chem. 134, 116133 (2021). https://doi.org/10.1016/j.trac.2020.116133.
(25) A. Moreira de Oliveira, C.A. Teixeira, and L.W. Hantao, Microchem. J. 172, 106978 (2022). https://doi.org/10.1016/j.microc.2021.106978.
(26) S.E. Prebihalo, A. Brockman, J. Cochran, and F.L. Dorman, J. Chromatogr. A 1419, 109–115 (2015). https://doi.org/10.1016/j.chroma.2015.09.080.
(27) N. Boegelsack, C. Sandau, D.W. McMartin, J.M. Withey, and G. O’Sullivan, J. Chromatogr. A 1635, 461717 (2021). https://doi.org/10.1016/j.chroma.2020.461717.
(28) J.V. Seeley, N.E. Schimmel, and S.K. Seeley, J. Chromatogr. A 1536, 6–15 (2018). https://doi.org/10.1016/j.chroma.2017.06.030.
(29) D.V. Gough, D.H. Song, S. Schöneich, S.E. Prebihalo, and R.E. Synovec, Anal. Chem. 91(11), 7328–7335 (2019). https://doi.org/10.1021/acs.analchem.9b01085.
(30) H. Cai and S.D. Stearns, J. Chromatogr. A 1669, 462930 (2022). https://doi.org/10.1016/j.chroma.2022.462930.
(31) B. Giocastro, M. Piparo, P.Q. Tranchida, and L. Mondello, J. Sep. Sci. 41(5), 1112–1117 (2018). https://doi.org/10.1002/jssc.201700824.
(32) A.M. Muscalu and T. Gorecki, Trends Anal. Chem. 106, 225–245 (2018). https://doi.org/10.1016/j.trac.2018.07.001.
(33) J.-F. Focant, A. Sjödin, and D.G. Patterson, J. Chromatogr. A 1040(2), 227–238 (2004). https://doi.org/10.1016/j.chroma.2004.04.003.
(34) K.L. Organtini, A.L. Myers, K.J. Jobst, J. Cochran, B. Ross, B. McCarry, E.J. Reiner, and F.L. Dorman, J. Chromatogr. A 1369, 138–146 (2014). https://doi.org/10.1016/j.chroma.2014.09.088.
(35) K. Tokita, K. Takazumi, T. Oshima, and T. Shigyo, J. Am. Soc. Brew. Chem. 72(2), 154–161 (2014). https://doi.org/10.1094/ASBCJ-2014-0402-01.
(36) M.S.S. Amaral and P.J. Marriott, Mol. Basel Switz. 24(11), E2080 (2019). https://doi.org/10.3390/molecules24112080.
(37) J. Fillion, F. Sauvé, and J. Selwyn, J. AOAC Int. 83(3), 698–713 (2000). https://doi.org/10.1093/jaoac/83.3.698.
(38) N. de A. Porto, J. Crucello, R. Facanali, I.M. Junior, R.M. Carvalho, and L.W. Hantao, J. Chromatogr. A 1655, 462485 (2021). https://doi.org/10.1016/j.chroma.2021.462485.
(39) G.C. da Silva, A.A.S. da Silva, A.L.S.N. da Silva, R.L. de O. Godoy, L.C. Nogueira, S.L. Quitério, and R.S.L. Raices, Food Chem. 167, 71–77 (2015). https://doi.org/10.1016/j.foodchem.2014.06.033.
(40) H.-Y. Shen and H.-L. Jiang, Anal. Chim. Acta 535(1), 33–41 (2005). https://doi.org/10.1016/j.aca.2004.12.027.
(41) “Standard Test Method for Sulfur Compounds in Light Petroleum Liquids by Gas Chromatography and Sulfur Selective Detection.” https://www.astm.org/d5623-19.html
(42) A. Arena, M. Zoccali, L. Mondello, and P.Q. Tranchida, Food Chem. 396, 133721 (2022). https://doi.org/10.1016/j.foodchem.2022.133721.
(43) A.M. de Oliveira, C.A. Teixeira, and L.W. Hantao, Anal. Methods 14(16), 1646–1654 (2022). https://doi.org/10.1039/D2AY00314G.
(44) A.M. Muscalu, E.J. Reiner, S.N. Liss, T. Chen, G. Ladwig, and D.A. Morse, Anal. Bioanal. Chem. 401(8), 2403–2413 (2011). https://doi.org/10.1007/s00216-011-5114-0.
(45) R.A. Shellie and P.J. Marriott, Analyst 128(7), 879–883 (2003). https://doi.org/10.1039/B304371A.
(46) K. Mastovská and S.J. Lehotay, J. Chromatogr. A 1000(1–2), 153–180 (2003). https://doi.org/10.1016/S0021-9673(03)00448-5.
(47) P.J. Marriott, S.-T. Chin, and Y. Nolvachai, J. Chromatogr. A 1636, 461788 (2021). https://doi.org/10.1016/j.chroma.2020.461788.
(48) N.D. Spadafora, S. Mascrez, L. McGregor, and G. Purcaro, Food Chem. 383, 132438 (2022). https://doi.org/10.1016/j.foodchem.2022.132438.
(49) P.E. Sudol, G.S. Ochoa, and R.E. Synovec, J. Chromatogr. A 1644, 462092 (2021). https://doi.org/10.1016/j.chroma.2021.462092.
(50) S.E. Reichenbach, X. Tian, C. Cordero, and Q. Tao, J. Chromatogr. A 1226, 140–148 (2012). https://doi.org/10.1016/j.chroma.2011.07.046.
(51) L.A. Adutwum, A.P. de la Mata, H.D. Bean, J.E. Hill, and J.J. Harynuk, Anal. Bioanal. Chem. 409(28), 6699–6708 (2017). https://doi.org/10.1007/s00216-017-0628-8.
(52) S.E. Prebihalo, G.S. Ochoa, K.L. Berrier, K.J. Skogerboe, K.L. Cameron, J.R. Trump, S.J. Svoboda, J.K. Wickiser, and R.E. Synovec, Anal. Chem. 92(23), 15526–15533 (2020). https://doi.org/10.1021/acs.analchem.0c03456.
(53) G.S. Ochoa, S.E. Prebihalo, B.C. Reaser, L.C. Marney, and R.E. Synovec, J. Chromatogr. A 1627, 461401 (2020). https://doi.org/10.1016/j.chroma.2020.461401.
(54) L.C. Marney, C.W. Siegler, B.A. Parsons, J.C. Hoggard, B.W. Wright, and R.E. Synovec, Talanta 115, 887–895 (2013). https://doi.org/10.1016/j.talanta.2013.06.038.
(55) M.S. Dunn, N. Vulic, R.A. Shellie, S. Whitehead, P. Morrison, and P.J. Marriott, J. Chromatogr. A 1130(1), 122–129 (2006). https://doi.org/10.1016/j.chroma.2006.08.012.
(56) European Chemicals Agency, “Compendium of Analytical Methods Recommended by the Forum to Check Compliance of REACH Annex XVII Restrictions:,” Publications Office: LU, 2021.
(57) US EPA, “O. EPA Method 8270E (SW-846): Semivolatile Organic Compounds by Gas Chromatography/Mass Spectrometry (GC–MS).” https://www.epa.gov/esam/epa-method-8270e-sw-846-semivolatile- organic-compounds-gas-chromatographymass-spectrometry-gc.
ABOUT THE AUTHOR
Sarah E. Prebihalo (pictured) is with the U.S. Food & Drug Administration, Center for Food Safety and Applied Nutrition, in College Park, Maryland. Brooke C. Reaser is with Agilent Technologies, Inc., in Santa Clara, California. Derrick V. Gough is with the U.S. Department of Defense at Rivanna Station, in Charlottesville, Virginia. Direct correspondence to: Sarah.Prebihalo@fda.hhs.gov


New Study Reviews Chromatography Methods for Flavonoid Analysis
April 21st 2025Flavonoids are widely used metabolites that carry out various functions in different industries, such as food and cosmetics. Detecting, separating, and quantifying them in fruit species can be a complicated process.
University of Rouen-Normandy Scientists Explore Eco-Friendly Sampling Approach for GC-HRMS
April 17th 2025Root exudates—substances secreted by living plant roots—are challenging to sample, as they are typically extracted using artificial devices and can vary widely in both quantity and composition across plant species.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)













