GSH Conjugate Metabolite Identification with a Hybrid Triple- Quadrupole–Linear Ion-Trap Instrument and Automated Software-Driven Method Development
Special Issues
In drug discovery, determining information about the extent of metabolism and the elucidation of metabolite structures is a vital step for lead optimization and drug scaffold refinement. The identification and characterization of metabolites plays an important role in both the drug discovery and development phases, as unsuitable pharmacokinetics (bioavailability and drug distribution), toxicity, and adverse drug reactions might be linked to metabolic instability. Historically, metabolite identification was carried out after a compound had been chosen for drug development. However, to reduce candidate failures attributed to toxicity effects, many pharmaceutical companies now conduct these experiments in the earliest phases of candidate drug selection.
The risk of idiosyncratic toxicity is of major concern to both pharmaceutical companies and regulatory bodies, especially because the low frequency of occurrence means that it can be missed by preclinical assessment and clinical trials. One possible cause of idiosyncratic toxicity is the formation of reactive metabolites, which are capable of covalent modification of proteins and nucleic acids via reactions such as nucleophilic substitution. It has been suggested that these types of reactions can contribute to idiosyncratic toxicity due to the interruption of certain cellular processes. While the process and causes of drug-induced idiosyncratic toxicity are not understood fully, the formation of reactive metabolites appears to be associated with various toxicological events. It is therefore desirable for pharmaceutical companies to screen for the formation of reactive metabolites.
Reactive–electrophilic metabolites are known to conjugate with the endogenous tripeptide, glutathione (γ-glutamyl-cysteinylglycine [GSH]), either spontaneously or through catalysis by GSH-S-transferases present in the cytosol and endoplasmic reticulum. Therefore, the presence of GSH-conjugated metabolites is an indication of the formation of reactive metabolites. As a result, it is widely accepted that the identification and characterization of GSH conjugates represents a valuable indirect approach for the identification of chemically reactive intermediates formed during the metabolism of both xenobiotic and endogenous compounds.
The complexity of different biological matrices combined with the low abundance of possible toxic GSH conjugates creates the need for highly selective and sensitive analytical tools for detection and identification of these types of metabolites.
MS-Based Approaches to Metabolite ID
With its sensitivity, selectivity, speed, and robustness, mass spectrometry (MS) coupled with high performance liquid chromatography (HPLC) has evolved into the preferred method for metabolite identification. Advances in software and hardware have resulted in a variety of techniques for tandem MS (MS-MS) that enable identification, quantitation, and characterization of many metabolites; however, many of the approaches are tedious and time-consuming.
Workflows commonly have consisted of sequential ion scans for metabolite identification followed by additional stages of MS for characterization, and extensive offline data analysis for confirmation. Different platforms offer varying attributes that are desirable to these tasks, but they can also possess drawbacks including limited sensitivity, throughput, structural information, and reliability.
Such strategies have included employing two MS systems — a triple-quadrupole system to find metabolites, via the benefit of fast selective scan functions such as precursor ion and neutral loss scans, and a separate ion-trap system to identify and characterize the metabolites, via the fast, high-sensitivity full scans that are possible on an ion-trap system. Samples would be split and run on the separate instruments sequentially, with the information generated on the triple-quadrupole system used to drive characterization on the ion-trap system. Other systems have relied on splitting the HPLC flow into two separate systems to generate information simultaneously. While the information in this type of configuration is acquired simultaneously, the data is generated into two separate files that must then be interrogated manually.
Hybrid Triple Quadrupole–Linear Ion-Trap MS
A hybrid triple quadrupole–linear ion-trap LC–MS-MS system (QTRAP system, Applied Biosystems, Foster City, California) (Figure 1) has been shown to be a powerful analytical tool for in vitro and in vivo metabolite detection and identification, including determining the presence of and characterization of GSH conjugates. The unique ability of this hybrid system to combine highly selective triple-quadrupole scan modes with fast, sensitive ion-trap MS-MS scans allows for optimal detection of drug metabolites as well as characterization and confirmation, in the same LC–MS run. The variety of survey modes (full-scan MS, precursor ion scans, neutral loss scans, and MRM), coupled with full-scan MS-MS confirmation via information-dependent acquisition (IDA), allows for a broad, sensitive, and effective system for metabolite detection.
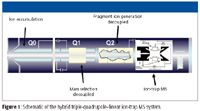
Figure 1
The triple-quadrupole scan modes on the hybrid system provide a selective method for identification of structurally similar metabolites even in the presence of complex biological backgrounds. Information based on the parent drug structure fragmentation can be utilized in the application of parent structure precursor ion scans and neutral-loss scans. A precursor ion scan essentially will find all of the masses in a sample that have the ability to lose a selected charged fragment related to the parent molecule. Neutral-loss scans select for those species that lose a characteristic uncharged fragment.
There are also several well-documented types of precursor and neutral-loss scan modes that can be applied specifically to certain types of phase II metabolites. Though there are several well-documented types of scan modes used, particularly for finding GSH metabolites, the most common are the neutral loss scan of 129 in positive ion mode and the precursor of m/z 272 scan in negative ion mode (Figure 2). Another well-known triple-quadrupole scan mode is multiple reaction monitoring (MRM). MRM is an extremely sensitive mode in which the desired precursor ion is selectively permitted into the collision cell (q2), and a specific product ion is filtered in the third quadrupole for confirmation and quantitation, if desired. A LINAC collision cell will permit the monitoring of up to 300 MRM transitions in a single experiment. MRM also has been shown to be excellent for GSH-conjugate detection, particularly when the Q3 transition is determined from the neutral loss of 129 from the expected conjugate Q3 mass.
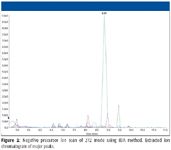
Figure 2
Ion-trap scan modes on a hybrid system provide the full-scan MS and MS-MS data often needed for metabolite identification. IDA mode is used most commonly, which allows data acquired in the survey scan (be it precursor, neutral loss, MRM, or full-scan MS) to select the ions for MS-MS acquisition "on the fly." In this manner, MS-MS data can be acquired automatically without the need to know parent drug information in advance.
Wen and colleagues (1) recently described a high-throughput approach to screening and characterization of reactive metabolites that takes advantage of both the unique hybrid system scan functions and the polarity switching capability of the instrument. Using the negative precursor ion scan to trigger the acquisition of positive enhanced product ion spectra, they developed a highly sensitive and efficient method for the detection and characterization of GSH-trapped reactive metabolites.
They found that by setting the negative precursor ion scan as the survey to monitor for the anion at m/z 272, they were able to detect a range of GHS conjugates. In their experiments, four of the seven clozapine GSH conjugates they detected with this method had not been reported previously in the literature. This unique screening strategy is extremely beneficial because the experiment design utilizes a selective negative precursor ion scan that is universal in finding GSH conjugates. It is followed by a positive ion full-scan MS-MS, enabling them to obtain structural information regarding GSH adducts, within a single run. An additional benefit they found with this new method was that they did not experience any false positives in human liver microsome (HLM) incubation.
Software Advances
Recently developed software (Lightsight 2.0 software, Applied Biosystems) enables easier method development through automation, allowing the creation of highly optimized methods much more quickly than was previously possible. The software can automatically construct optimized methods specifically for GSH-conjugate screening and characterization. These include methods utilizing GSH-specific survey scans such as neutral loss of m/z 129 and precursors of m/z 272, emulating the workflow demonstrated by Wen and colleagues.
As an alternative method of metabolite detection and identification, the software can create compound-specific predictive MRM-IDA methods (pMRM-IDA), increasing metabolite detection over traditional strategies. With this approach, with prior knowledge of the parent drug and its fragmentation pathway, an onboard method creation tool determines the theoretical MRM transitions for metabolites based upon a comprehensive biotransformation set derived from the literature. One advantage of the pMRM methods generated by the Lightsight 2.0 software, over previous methods, is that the software better predicts MRM transitions for metabolites that are a result of a combination of modifications. The software also can generate a complete set of MRM transitions for metabolites that are result of three or four modifications, giving rise to multiple permutations of MRM transitions.
Experimental Example
To demonstrate the utility of the combination of the hybrid system with method development software in detecting GSH conjugates, an in vitro incubation of troglitazone in rat hepatocytes at a concentration of 10 μM was carried out with time points at zero and four hours. Samples were analyzed using a 4000 QTRAP hybrid triple-quadrupole–linear ion-trap mass spectrometer (Applied Biosystems). Scans used were the neutral loss scan of 129 and precursor ion scan of m/z 272, with optimum conditions determined by Lightsight software (Figures 3 and 4).

Figure 3
A total of 10 GSH conjugates were found in the in vitro incubation of troglitazone using a combination of the two different scan modes. The negative mode precursor scans appear to be more effective at detecting GSH conjugates than the positive mode neutral loss scan, despite the positive mode neutral loss generally having a higher signal-to-noise ratio. This might be because the positive mode neutral loss 129 generates more false-positive peaks than the negative mode precursor scans, thereby making the detection of true GSH conjugates more difficult.
Both positive and negative MS-MS of GSH conjugates are useful for this type of analysis. Negative mode MS-MS usually predominantly comprises GSH fragments, and therefore can be used to confirm the presence of a GSH metabolite, while positive mode MS-MS often yields structural information about the GSH conjugate. Therefore, the software automatically couples the negative mode precursor of m/z 272 survey scan with a positive ion MS-MS IDA dependent scan. This combination of negative ion precursor survey scan and positive ion dependent scan allows for the more effective detection of GSH conjugates of the precursor scan and the structural information derived from the positive ion MS-MS IDA scan.
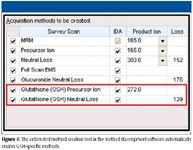
Figure 4
Conclusion
In today's frenetic world of pharmaceutical development with an ever-expanding base of new candidate compounds and an ever-increasing vigilance for safety, metabolite identification and characterization is an even more critical component of the drug discovery process. The examples presented demonstrate the benefit of polarity switching between MS detection and MS-MS acquisition for rapid screening and characterization of reactive metabolites. Compared with the typical neutral loss methods, negative ion mode analysis of the precursors of m/z 272 demonstrates several advantages. First, the superior selectivity identifies many different classes of GSH conjugates with essentially no false positives. Second, the sensitivity is improved significantly for detecting minor GSH conjugates at low levels. Finally, method development software makes the setup, acquisition, and data reduction straightforward, and the hybrid triple-quadrupole–linear ion-trap system is highly efficient in the detection and structural characterization of GSH conjugates all in a single LC–MS-MS run.
The latest generation of MS-based tools are more accurate and sensitive, enable higher throughput for laboratories, and require less specialized training for investigators to generate thorough results. A hybrid MS system enables its operators to consolidate identification and confirmation experiments that might otherwise be necessary to conduct step-wise.
Acknowledgment
The authors would like to thank our collaborators who contributed to the information presented in this article: Ji Ma & Robert Cho Amgen, South San Francisco, California; Elliott Jones and Hua-Fen Liu, Applied Biosystems, Foster City, California.
Claire Bramwell is with Applied Biosystems, Foster City, California. Julie Wingate is with Applied Biosystems, Concord, Ontario. Christopher Elicone is with Applied Biosystems, Framingham, Massachusetts.
References
(1) B. Wen, L. Ma, S.D. Nelson, and M. Zhu, Anal. Chem. 80(5), 1788–1799 (2008).

New TRC Facility Accelerates Innovation and Delivery
April 25th 2025We’ve expanded our capabilities with a state-of-the-art, 200,000 sq ft TRC facility in Toronto, completed in 2024 and staffed by over 100 PhD- and MSc-level scientists. This investment enables the development of more innovative compounds, a broader catalogue and custom offering, and streamlined operations for faster delivery. • Our extensive range of over 100,000 high-quality research chemicals—including APIs, metabolites, and impurities in both native and stable isotope-labelled forms—provides essential tools for uncovering molecular disease mechanisms and exploring new opportunities for therapeutic intervention.
New Guide: Characterising Impurity Standards – What Defines “Good Enough?”
April 25th 2025Impurity reference standards (IRSs) are essential for accurately identifying and quantifying impurities in pharmaceutical development and manufacturing. Yet, with limited regulatory guidance on how much characterisation is truly required for different applications, selecting the right standard can be challenging. To help, LGC has developed a new interactive multimedia guide, packed with expert insights to support your decision-making and give you greater confidence when choosing the right IRS for your specific needs.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)










