Calibration Stability for Formula Determination on a Single-Quadrupole GC–MS System
Special Issues
With electron ionization (EI) used in most gas chromatography–mass spectrometry (GC–MS) applications, the molecular ion typically is broken apart into multiple fragment ions during the EI process.
With electron ionization (EI) used in most gas chromatography–mass spectrometry (GC–MS) applications, the molecular ion typically is broken apart into multiple fragment ions during the EI process. These multiple EI fragments complement each other to enhance the compound identification capability greatly through the use of MS library search (for example, NIST MS library search), in spite of the fact that the mass-to-charge ratio (m/z) measurement from the sample and all m/z values in the library are accurate only to about ×0.5 Da. Consequently, EI library search has become the method of choice for GC–MS compound identification. While this works well for many routine applications, ambiguity in compound identification can occur in the case in which similarly high search scores are obtained, corresponding to multiple compounds. In demanding applications such as the identification of impurities, degradants, and active ingredients in pharmaceutical, chemical, food, flavor, and fragrances industries, these unknown compounds might not even be in a database to start with. Consequently, these applications would require accurate mass measurement from a higher-resolution MS system such as GC–time of flight (TOF), which is more expensive and requires higher maintenance. The accurate mass measurement would serve either to confirm the library search results or to determine the elemental compositions of molecular ions and key fragments.
In a separate publication, Chen and colleagues (1) reported that it is entirely feasible to perform the same highly accurate measurements on a conventional quadrupole GC–MS system, through the use of a novel calibration scheme to calibrate both m/z and mass spectral instrument line shape elaborately. Because this elaborate calibration scheme involves both mass accuracy and spectral accuracy, a question typically arises concerning the stability of such an extensive calibration and its implications for practical applications.
This article reports on the design and results of a short-term calibration stability study in terms of both mass accuracy and spectral accuracy, and their impacts on elemental composition determination using a conventional single-quadrupole GC–MS system.
Theory
The conventional mass spectral calibration determines only the mass (m/z) alone without any consideration of the instrument line shape, which can impact the mass measurement significantly, especially on a unit mass resolution instrument such as a quadrupole GC–MS system. Instead, a novel calibration approach has been developed (2,3) that not only calibrates the mass axis, but also calibrates the mass spectral instrument line shape. This is uniquely possible in MS because the theoretical peak positions are known very accurately (they are simply the masses of corresponding ions). In addition, we also know quite accurately the various isotopes and their relative abundances. This information allows the calculation of a calibration function that corrects not only the peak position, but the instrument line shape as well.
With this new calibration technique, the actual instrument line shape, typically unknown and undefined, is calibrated into a symmetrical and mathematically defined line shape function. This can improve all downstream processing dramatically, including accurate monoisotopic mass determination to achieve high mass accuracy on a single-quadrupole GC–MS instrument. Conventionally, with just accurate mass information, the elemental composition has been determined by searching for all chemically possible formulas within a mass tolerance window from the reported accurate mass. Depending upon the inherent mass accuracy of the instrument, all formulas found within the mass tolerance must be considered. For example, for an instrument capable of a mass accuracy of 1 ppm, 34 formula candidates for a compound at 500 Da bounded by the elements C, H, N, O, S, and Cl are identified.
It is known that every unique formula has a unique isotope distribution, and that, in theory, could be used as additional information for formula identification. Previously, however, such methods were not considered viable for formula identification because it was difficult to model the isotope patterns on any given instrument to a spectral accuracy of better than 98% (4). This has limited the use of isotope profile information to assist in reducing the number of formula candidates generated from accurate mass measurements. If, however, the spectral accuracy could be improved to better than 99%, it could be used to identify a unique formula from a list of possible formulas generated by the accurate mass measurement, or even provide confident formula identification on instruments that are not calibrated as frequently, as long as similarly high spectral accuracy can be ensured.
With the calibrated instrument line shape function described earlier, a theoretical mass spectrum can be calculated exactly for each candidate formula from the list and compared with the measured mass spectrum after the previous calibration. A spectral accuracy metric of better than 99% can then be calculated to reflect any minor yet elementally significant difference between the two and rank the likelihood of a given formula being the correct answer. This process is called calibrated line-shape isotope profile search (CLIPS) and has been demonstrated to be more informative than mass accuracy for formula determination (1).
Experimental
Perfluorotributylamine (PFTBA) calibration standard and a sample mixture of compounds were measured on an Agilent GC–MS single-quadrupole MS 5973 system (Santa Clara, California) over a one-week period. The PFTBA standard was measured once through infusion and repeated again a week later at the end of the experimentation. The mixture sample was analyzed 22 times at different concentration levels between the two PFTBA runs, in which one blind test ion (nominal mass at m/z 162) was to be identified for its elemental composition using MassWorks CLIPS (Cerno Bioscience, Danbury, Connecticut). Both the PFTBA and the sample mixture were acquired under the same MS conditions in "raw" mode (no-peak detected, sometimes called profile mode) over a mass range of 50–550 m/z with ion threshold set to zero.
The same calibration, created with eight known fragments from the first PFTBA run, was applied to all 22 GC–MS runs to report the accurate masses of this blind test ion followed by its elemental composition determination. This same calibration also was applied to the second PFTBA run to report the accurate masses of the same eight known fragments in order to measure the mass drift between these two PFTBA runs.
Results and Discussion
Figure 1 shows the total ion chromatogram of the first PFTBA run and the 0.8-min retention time window used for the mass spectral averaging before the calibration. Figure 2 shows the profile mode mass spectrum before and after the calibration for the calibration ion C3F5+ . This demonstrates that the raw profile mode mass spectrum, although very well tuned, does not have a known or defined peak shape, whereas the calibrated mass spectrum does have a known and mathematically defined peak shape. The calibrated mass spectrum also has been shifted properly to the correct mass position given by the monoisotopic mass of this calibration ion. As a convention and without the loss of generality, the mass difference due to the gain or loss of an electron is ignored throughout this article and the exact mass corresponds to that of the neutral form of the same ion.
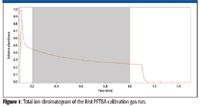
Figure 1
This calibration was applied to the second PFTBA run acquired one week later, and the reported accurate masses for the same set of eight calibration ions are listed in Table I. We see a systematic mass shift of about +7 mDa across all of these ions. When measured in parts per million (ppm), such mass shift amounts to +12 to +54 ppm mass error, typically deemed too large for elemental composition determination through the conventional monoisotopic mass search.

Figure 2
The same calibration was applied to the 22 GC–MS runs acquired within the week, and the accurate masses for the m/z 162 ion are plotted in Figure 3 along with its exact mass, calculated after the identity of the blind test ion was revealed to be nicotine (C10H14N2) and its ion signal counts. There is a clear correlation between the mass error and the ion signal level, as expected and established in the literature (5). For intensities above 8000 counts, all reported accurate masses come within a few millidaltons of the exact mass. This is consistent with the overall mass shift estimate from Table I and confirms that the overall mass shift observed between the two PFTBA runs are more or less comparable to the random fluctuation of the repeated mass measurement at similar ion intensities. In other words, the overall systematic mass shift error of ~7 mDa is not statistically significant when compared with the random fluctuation of the mass measurement itself (that is, the first PFTBA calibration is indeed applicable to all 22 GC-MS runs acquired afterward without causing significant systematic mass measurement errors).
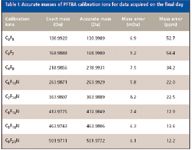
Table I: Accurate masses of PFTBA calibration ions for data acquired on the final day
As has been reported (6), while mass accuracy is important for formula determination, it is typically just a single number picked out of the complete isotope response of a given ion. For this reason, there are many formula candidates whose exact monoisotopic masses fall within a small mass tolerance window of a reported accurate mass, even when measuring with a high-resolution mass spectrometer with 1–2 ppm mass accuracy. Spectral accuracy, an independent metric, measures how well an actually measured mass spectrum matches that obtained from theoretical calculation. It contains much more information about the fine structures of the isotope distribution for a given ion and might be more important than mass accuracy. Furthermore, the spectral accuracy metric might be more robust than mass accuracy for the purpose of formula determination, since the mass spectral instrument line shape on which spectral accuracy depends might be less susceptible to time-related variations on a mass spectrometer.

Figure 3
The unknown ion at 162 Da with its mass error of more than 50 ppm (Table I and Figure 3) offers a good example to test the relative importance and robustness of mass accuracy versus spectral accuracy. Treating the 162 Da ion as a total unknown with common elements C, H, N, O, F, and Cl included under general elemental composition search conditions (Table II), all 22 mass spectral measurements of this same ion were run through the software to determine its formula. Ranked by spectral accuracy, the formula C10H14N2 came as the first hit 19 times, as the second hit two times, and as the third hit once (Figure 4). This formula was later confirmed to be the correct formula for nicotine. The second hit and third hit all correspond to lower concentration samples (Figure 3).

Table II: MassWorks CLIPS formula determination parameters
Take three runs: 24–17–12, 26–18–29, and 31–11–40 as examples and overlay the theoretically calculated mass spectrum for nicotine on top of the software calibrated version. We see from Figure 5 that a good quality spectral match is attained for the lower concentration run (24-17-12) and a near perfect spectral match is attained for the higher concentration runs (26-18-29 and 31-11-40), all reflected in their corresponding spectral accuracy metric. Figure 5 offers evidence that the spectral accuracy is directly related to ion signal level (noise in data) with little or no discernable correlation to time or mass shift. This indicates that spectral accuracy is not only more informative but also far more robust for elemental composition determination.
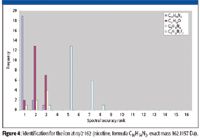
Figure 4
Figure 5 depicts a section of the mass spectrum corresponding to the 162 Da nicotine ion. This shows a mass spectral peak at 161 Da with about the same ion intensity as the nicotine ion itself. As often happens in GC–MS and tandem MS-MS experiments, there might be other ions or fragments formed during the ionization–fragmentation process that lead to such isotope response overlaps. This mass spectral interference has caused some systematic mass measurement errors for the 162 Da ion, as evidenced by the mostly negative mass measurement bias from Figure 3. This does not affect the spectral accuracy calculation because the 161 Da ion also is added to the CLIPS calculation by modeling it as an M-H ion, where M is the unknown 162 Da ion for which the elemental composition is to be determined. In theory, many more interference ions similar to the 161 Da ion could be handled, though only up to two interferences can be included at this time.

Figure 5
Although the single-quadrupole GC–MS system used here has mass variation on the order of 7 mDa or more than 50 ppm at 162 Da during the one-week test period, an error large enough to cause alarms for conventional accurate mass elemental composition determination, the mass spectral line shape reflected by the spectral accuracy metric remained unchanged throughout the week, leading to consistently high confidence formula determination without recalibration.
Conclusion
This short-term stability study shows that the elaborate calibration involving both mass accuracy and spectral accuracy does not render the calibration more susceptible to time-dependent variations. Furthermore, the formula determination approach maintains its robust performance through the separation of mass accuracy and spectral accuracy. While the mass accuracy fluctuated by about 7 mDa during the one-week test period, the mass spectral instrument line shape or spectral accuracy experienced no noticeable changes, allowing for robust formula determination without recalibration. This is achieved by taking advantage of the exact isotope modeling process embedded in the CLIPS algorithm, which fully exploits the fine structures in the whole isotope distribution of a given ion instead of relying on a single accurate mass reading from its monoisotopic peak.
Acknowledgment
The authors would like to thank Ms. Alice Laures and her colleague Mr. Ian Collin from GlaxoSmithKline (Brentford, Middlesex, United Kingdom) for acquiring the GC–MS data used in this study.
Ming Gu and Yongdong Wang are with Cerno Bioscience, Danbury, Connecticut.
References
(1) J.P. Chen et al., LCGC, in press.
(2) M. Gu et al., Rapid Commun. Mass Spectrom. 20, 764–770 (2006).
(3) Y. Wang, Methods for operating mass spectrometry (MS) instrument systems, US Patent 6,983,213, filed October 20, 2003.
(4) T. Kind, BMC Bioinformatics 2006, 7, 234 (www.biomedcentral.com/1471-2105/7/234).
(5) K.R. Blom, Anal. Chem. 73, 715 (2001).
(6) D. Kuehl, Am. Lab Online Edition, January 2008, Page 18, americanlaboratory.texterity.com/al-online/200801ol/?pg=18

Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.
Extracting Estrogenic Hormones Using Rotating Disk and Modified Clays
April 14th 2025University of Caldas and University of Chile researchers extracted estrogenic hormones from wastewater samples using rotating disk sorption extraction. After extraction, the concentrated analytes were measured using liquid chromatography coupled with photodiode array detection (HPLC-PDA).











