Detecting Pesticides in an Apple Matrix Using GC?GC–TOF–MS Without Complex Sample Preparation
The Application Notebook
ALMSCO Application Note
Introduction
Quantitative analysis of toxic organic compounds in complex food matrices is a challenging problem, especially if both library compounds and unknown components are to be identified in the same chromatographic run. Furthermore, analysts often have to resort to complex sample preparation to avoid background interference, which can complicate the identification and quantification of analytes.
Here, we demonstrate that GC×GC and time-of-flight (TOF)-MS can detect ppb levels of pesticides in a complex apple extract, with minimal sample preparation.
The TOF instrument used, ALMSCO's BenchTOF-dx, is designed fundamentally for sensitivity, but also has good mass accuracy. This enables it to detect target compounds currently confirmed by triple-quadrupole (QQQ) instruments, to similar levels and in similar food matrices. Not only that, but because of its data acquistion rate of 10000 spectra per second, the output of the BenchTOF-dx can be compared against standard libraries. This enables fast analysis of all the components in the mixture.
Experimental
The extraction technique used was chosen because it was both quick and cheap (as well as being used by many regulated European labs).
A sample of an organic Braeburn apple (60 g) was cut into pieces, added to ethyl acetate (60 mL), and the mixture agitated for 10 min. The organic layer was then separated, dried over sodium sulphate, filtered, and spiked to 10 ppb with a mix of pesticides and PCBs. This was then brought down to 100 ppt by dilution using more apple matrix. Mirex was spiked into all calibration levels at 2.5 ppb.
For details of the GC×GC and TOF-MS conditions, please see the application note ANBT15 (available at http://bit.ly/ALMSCOapps).
The program GC Image (Zoex, Inc.) was used for visualisation and data processing.
Results and Discussion
Although standard GC and TOF-MS gives good results for this sample, comprehensive GC×GC is an even better solution for full analysis of such a complex mixture. This involves five-second aliquots being taken from the first column and run through the second column, with the results displayed in two dimensions, as shown in Figure 1.

Figure 1: Total ion chromatogram of the apple extract, including a 2.5 ppb Mirex internal standard (the narrow peak in the centre of the chromatogram).
However, even with this level of separation, matrix compounds dominate the chromatographic space. To avoid this, the data was reprocessed, to pick out just those peaks containing ions close to the mass of Mirex (271.81 ± 0.05 Da). In this so-called extractedion chromatogram, the matrix interferences have been completely eliminated (Figure 2).

Figure 2: A 0.1 Da extracted-ion chromatogram of the apple extract, showing just the Mirex signal.
A key benefit of the spectra produced by the BenchTOFdx is the high degree of matching with the NIST database. Figure 3 compares the mass spectrum of the Mirex peak with that of the reference spectrum in the NIST05 database. Not only are the forward and reverse match coefficients very high, but the 98% probability score removes any ambiguity about the assignment.
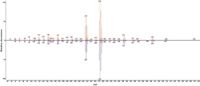
Figure 3: BenchTOF-dx mass spectrum (top) of the Mirex peak, compared with that of a reference NIST05 mass spectrum (bottom). The forward and reverse match coefficients are 894 and 905, respectively.
Finally, to demonstrate the power of this analytical method to handle a wide range of analyte concentrations, calibration curves were plotted for two representative components of the mixture, 3-chlorobiphenyl and 4,4'-DDD (Figure 4), with excellent linearity being observed.
Conclusions
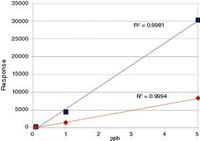
Figure 4: External calibration curves plotted over the concentration range 100 ppt to 5 ppb for 3-chlorobiphenyl (blue squares) and 4,4â²-DDD (red diamonds).
This work demonstrates that the enormous selectivity of comprehensive GC×GC, combined with the sub-unit-mass resolving power of the BenchTOF-dx, enables detection of trace-level target analytes in apple extract.
Importantly, the method uses a very simple extraction method, making it a strong candidate for the routine screening of persistent organic pollutants in complex food matrices.
ALMSCO International
Gwaun Elai Medi Science Campus, Llantrisant, RCT CF72 8XL, UK
tel: +44 (0)1443 233920 fax: +44 (0)1443 231531
E-mail: enquiries@almsco.com Website: www.almsco.com

Analyzing Vitamin K1 Levels in Vegetables Eaten by Warfarin Patients Using HPLC UV–vis
April 9th 2025Research conducted by the Universitas Padjadjaran (Sumedang, Indonesia) focused on the measurement of vitamin K1 in various vegetables (specifically lettuce, cabbage, napa cabbage, and spinach) that were ingested by patients using warfarin. High performance liquid chromatography (HPLC) equipped with an ultraviolet detector set at 245 nm was used as the analytical technique.
Removing Double-Stranded RNA Impurities Using Chromatography
April 8th 2025Researchers from Agency for Science, Technology and Research in Singapore recently published a review article exploring how chromatography can be used to remove double-stranded RNA impurities during mRNA therapeutics production.
The Effect of Time and Tide On PFAS Concentrations in Estuaries
April 8th 2025Oliver Jones and Navneet Singh from RMIT University, Melbourne, Australia discuss a recent study they conducted to investigate the relationship between tidal cycles and PFAS concentrations in estuarine systems, and offer practical advice on the sample preparation and LC–MS/MS techniques they used to achieve the best results.
Assessing Safety of Medications During Pregnancy and Breastfeeding with LC–MS/MS
Published: April 8th 2025 | Updated: April 8th 2025Researchers conducted a study on medications used in psychiatry and neurology during the perinatal period, assessing how antiepileptic drugs (AEDs) affect placental functions, including transport mechanisms, nutrient transport, and trophoblast differentiation. Several quantitative methods, such as those for antianxiety and hypnotic drugs, were established to evaluate the safety of pharmacotherapy during breastfeeding using liquid chromatography-tandem mass spectrometry (LC–MS/MS).




















