Challenges in Small-Molecule Quantitation by Mass Spectrometry
Special Issues
Drug discovery scientists are continually striving to improve productivity and efficiency in their workflows. From early discovery to clinical development, existing workflow bottlenecks represent an opportunity to develop solutions to speed the process and improve productivity. The key requirements for quantitative analysis are precision, accuracy, and linear dynamic range. With any quantitative instrument, the hope is that it will be applicable to a vast range of coumpounds, ruggest, and fast. New mass spectrometry (MS) technologies are being developed that meet these criteria and permit high throughput while enabling its application to areas in which speed limitations previously curtailed its practicality. In particular, in the area of ADME profiling, new MS platforms are becoming available that increase the throughput by at least 25-fold, by combining the speed of matrix-assisted laser desorption ionization (MALDI) with the specificity of triple-quadrupole MS. This is bound to greatly accelerate the ADME..
Drug discovery and development scientists are continually pressed to increase productivity and efficiency in their workflows, and the area of absorption, distribution, metabolism, and excretion (ADME) profiling is no exception. The typical ADME laboratory is tasked with routinely screening and profiling hundreds of compounds via a variety of assays to assess their suitability as potential drugs. Each compound generates multiple samples, creating the need for very high throughput methods for handling the volume of testing in the laboratory. Although these types of in vitro assays typically do not require high sensitivity or a wide dynamic range, speed and reproducibility are of paramount importance. However, the liquid chromatography (LC) bottleneck and analysis time of up to several minutes per sample limit the number of compounds that can be profiled by mass spectrometry (MS) and, as critically, the turnaround of profiling results. Current approaches to reducing the LC–MS-MS bottleneck include running parallel systems to maximize analytical time capacity and high-pressure systems to increase analysis pace, but these alternatives increase both costs and complexity without offering a definitive solution to the throughput issue.
Upstream in the drug discovery pipeline, the advent of high-throughput screening (HTS) technologies using fluorescence and chemiluminescence-based assays combined with advancements in robotics and liquid handling have enabled screening studies on libraries containing up to millions of compounds. While the high capacity provided by these assays is highly desirable, the reagents used to generate the fluorescence or chemiluminescence read-out signal can interfere with certain compounds, creating false-positive or false-negative results and thus leading to incorrect or ambiguous conclusions. In addition, these reagents are relatively costly. While MS is recognized as among the most specific technologies available to resolve those ambiguities, it was not possible to match the fast screening capabilities of current assays on an MS platform, and this approach currently is limited to occasional confirmation studies on small subsets of compounds.
New Solutions
A new solution to these challenges is the development of new MS platforms and software that combine the speed of matrix-assisted laser desorption ionization (MALDI) with the specificity of triple-quadrupole (QqQ) MS to increase the analytical throughput by at least 25-fold. This can greatly accelerate the ADME profiling of lead compounds and increase operational flexibility by decoupling sample preparation from the MS analysis (Figure 1).
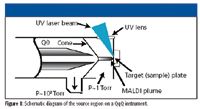
Figure 1
In developing a MALDI QqQ system, engineers needed to keep in mind the key requirements for small molecule quantitative analysis in biological samples; tandem MS is needed for selectivity, a high repetition rate laser provides sensitivity and speed, sample cleanup is required for proper crystallization and reduction of ion suppression, and an internal standard is necessary for precision (1). Each of these were considered when evaluating the technology for discovery ADME application workflows as they pertain to compound coverage, sensitivity, linear dynamic range, precision and accuracy, and overall speed and throughput of the technique.
The use of MALDI over the years primarily has been for the analysis of large, intact molecules (peptides and proteins) on time-of-flight (TOF) technologies. TOF detectors are considered less suited for quantification because the dynamic range for a quantitative assay is generally about two orders of magnitude.
The usefulness of MALDI for small molecule work was limited by several factors, including the MALDI matrix itself, which results in significant interference in the low mass range of a mass spectrum <500 Da, sample preparation, and various crystallization and sample deposition issues.
Recently, however, its use has seen an emergence in small molecule analysis for high-throughput screening applications in discovery ADME workflows. High-repetition lasers have been incorporated to improve throughput and sensitivity. Efficient ionization of small molecules by MALDI is accomplished in the presence of α-cyano hydroxycinnamic acid (CHCA) (2). Peptides also can be ionized very efficiently in the same conditions, being limited only by the upper limit of the mass range of the quadrupole mass spectrometer used in conjunction with the MALDI source (2800 m/z for the 4000 QTRAP system; 3000 m/z for the API 4000 system [Applied Biosystems, Foster City, California]). This capability enables the monitoring of small peptide substrate–product pairs used in many enzyme inhibition assays.
With the specificity and sensitivity of multiple reaction monitoring (MRM) on a triple-quadrupole mass spectrometer and the throughput advantages of MALDI ionization, the combination provides a significant improvement in overall throughput, where the speed of MALDI QqQ analysis can reach 1 s/sample with adequate sensitivity, accuracy, and precision for in vitro ADME quantification (3). This is on the order of 25 to 40 times faster than the most efficient LC–MS-MS methodologies, making it very attractive to analytical laboratories that need to boost productivity and efficiency. The system has the capability to go faster (2–3 samples/s) for HTS applications, in which precision is less critical.
Specificity
Triple-quadrupole instruments can achieve orders of magnitude in specificity when operated in MRM mode (selection of specific parent-to-daughter ion transitions). The high signal-to-noise ratio (S/N) obtained by using tandem MS allows for limits of detection (LOD) that are more than adequate for most ADME applications. Being able to collect MRM data at very high speeds becomes a significant advantage of MALDI source technology on a triple quadrupole instrument. For assays requiring lower limits of detection and for those compounds that do not ionize by MALDI, electrospray ionization (ESI) MS-MS is the complementary technology. The high-repetition laser source can be interchanged with an ESI source within approximately 15 min, excluding instrument pump down time.
In the MALDI QqQ platform, samples for analysis are deposited directly onto a MALDI target surface after precleaning; standard 96 or 384 plate formats are compatible with the system. Several different rastering modes are available to ablate the sample spot and generate an analyte peak, but in general, straight-line raster is recommended to achieve fast quantitative analysis.
Figure 2a shows an example of a straight line raster across a sample spot, and Figure 2b shows the corresponding analyte peak detected for the MRM transition monitored.
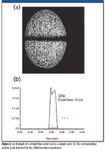
Figure 2
Application Examples
As previously mentioned, while sensitivity and linear dynamic range requirements for early discovery ADME applications typically are not demanding, speed and overall throughput are of utmost importance. Several applications fitting these criteria were tested, including metabolic stability screening, cytochrome P450 (CYP) inhibition, and permeability assays.
For example, hepatic clearance is an important pharmacokinetic property of a drug candidate. High clearance causes poor oral bioavailability due to first-pass metabolism, and a short half-life. Because of the key importance of this property, new compounds routinely are evaluated at the early discovery stage using in vitro models of hepatic metabolism, such as metabolic stability profiling in liver microsomes and hepatocytes.
Microsomal incubation in rate liver microsomes was performed on buspirone at 0-, 5-, 15-, and 30-min time points in a 96-well incubation format. The resulting samples were transferred into two 96-well plates, one for MALDI–MS-MS and the other for LC–MS-MS comparison. A solid-phase extraction cleanup was performed on the MALDI samples before spotting a 1-μL aliquot of sample–matrix mix to the plate. Figure 3 shows a typical metabolic stability profile for this analysis with high correlation between the two techniques. The key difference with the MALDI-MS-MS data set is that the entire 96 sample plate was complete in 5 min as compared with an LC–MS-MS data set that required 1.6 h for the same number of samples.
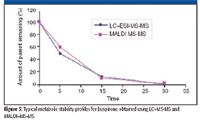
Figure 3
Drug–drug interactions are another typical screen routinely run in early ADME. Potent inhibition of CYP enzymes is a major cause of drug-induced toxicity. Drug candidates are routinely screened at a very early stage for their potential to inhibit various CYP isoforms using in vitro assays. Inhibition of major isoforms such as CYP 3A4 and 2D6 is considered a major liability and often causes the failure of a drug candidate. While the majority of inhibition screening is done using non-MS-based assays such as fluorescence, certain key isoforms do not have a suitable fluorescent substrate. LC–MS-MS is used for such nonfluorescent substrates. Because these assays typically are performed in a high-speed automated format, LC–MS-MS analysis is a bottleneck in this workflow.
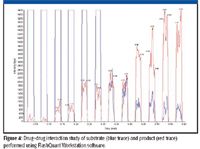
Figure 4
Software for MALDI-MRM
Eliminating a bottleneck from one part of a process usually uncovers a new bottleneck elsewhere. For the drastically increased speed of sample data acquisition on a MALDI QqQ platform, a complete software program for setting up and running 96- or 384-spot MALDI plates for the quantitation of small molecules in a nonregulated discovery environment was developed. The functions provided allow the user to complete the quantitation task at an appropriately accelerated pace.
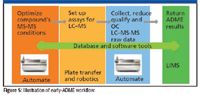
Figure 5
The methods used in designing and developing the FlashQuant software (Applied Biosystems) were selected specifically to ensure that even a new technology would closely meet the user's needs. To ensure that the application met users' needs as closely as possible, rapid prototyping, extreme programming (4), and usability testing (5) were key methods used, with many software functions added or adjusted based upon user feedback before completion. The additional effort for these methods was found to be worthwhile in meeting the objective.
A single integrated workspace displays all the data associated with the targeted analytes of interest. For a typical MALDI-MRM quantitation workflow, the software allows the user to obtain quantitative information from their small molecule quantitative application. From the MALDI source control (including laser power, frequency, x–y stage movement, and gas flows), manual and batch acquisition, to MRM quantitative data processing and reporting, the workstation simplifies the MALDI-MRM quantitation experiment from sample analysis to report in three steps.
New Software Approach for LC–MS Systems
To support existing LC–MS analytical workflows, another new software application (DiscoveryQuant, Applied Biosystems) enables higher throughput quantitation in the early discovery laboratory. Software with automated tools, shared information, and automated assay setup can improve the ability of the ADME laboratory to broaden its pharmaceutical pipeline by gaining more information on a higher number of potential compounds.
While earlier high-throughput software programs for ADME offered automated method development, acquisition, and data processing of target compounds, they also had limitations. Accessible primarily for advanced users, programs were challenging to initiate and relatively inflexible.
One key advantage of being able to perform proactive optimizations on hundreds of compounds a week is the ability to use them later, potentially with other mass spectrometers in other laboratories. With high-throughput discovery software, each optimized set of conditions can be stored in a networked database and recalled for use in assays at any time in the future, in any location. As the pharmaceutical enterprise data environment becomes globally connected, the ability for users to take advantage of the conditions-optimization work done previously, creating part of an analytical compound resume for each compound in the organization, is quite useful. Moreover, being able to exchange optimized compound parameters between MALDI and ESI platforms, the analyst now has the new possibility of selecting the optimal ionization technique to achieve the right combination of speed, sensitivity, and precision required for a particular assay. This added flexibility also maximizes compound coverage, because chances of encountering a poor ionizer on the two types of ion sources are reduced significantly.
Discovery laboratories generally have a set of routine assays that they perform. These assays tend to be set up in the same plate format each time they go through the laboratory, regardless of the compound. High-throughput discovery software allows laboratories to exploit this phenomenon by creating an assay template for standard assay types. The template lays out the samples on the plate so that users need only choose the compounds on the plates (from the compound database) and the type of assay template. The methods for each well on each plate are then automatically constructed for submission to the mass spectrometer.
A typical drug trial work involves large numbers of samples with the same analyte and internal standard. It tends to focus on excellence in quantitation and sensitivity for a single compound or set of compounds that are measured repeatedly. Mass spectrometers tend to excel at this workflow: once a method is optimized, it can be used again as often as needed. The variables are multiple subjects, multiple time-points, and multiple models, all looking for the same analytes.
Early-ADME work, however, is one of the few fields in which a vast variety of new compounds are continually being manufactured. Analysis needs are more akin to a simple "filtering" of each compound, and therefore, optimization of conditions cannot be a custom process.
Expanding out the workflow for early-ADME, there are actually four key stages to performing truly high-throughput screening. Being able to optimize and store the MS-MS conditions is critical. The second stage is the ability to set up plates of assays for the large numbers of new chemical entities quickly. This relies on robotics, which differ from laboratory to laboratory, and the techniques used to program them to reformat the compounds into the standard assay vials. Once this is done, and after the assay is complete, these need to be quantitated with LC–MS, using the optimized conditions in the first stage but producing an order of magnitude more data. This must then be checked for accuracy and precision before being sent as a completed ADME result to the storage system.
Discussion
The ongoing need for increased productivity, throughput, and speed in discovery ADME applications will continue to drive developments in MS technology and workflows. Emerging new MALDI QqQ MS systems and user-focused software applications are a viable option for dramatically improved sample throughput. Additional improvements will be made in time to hasten sample preparation, and in the understanding of its effect on MALDI quantitation, that will continually improve throughput.
For a high-throughput ADME setting, and potentially for many other high-throughput analytical laboratories, the tremendous increase in capacity made available with these solutions could deliver myriad benefits, most obviously, the ability to screen more leads through a standard set of assays, generating more data with the same level of resources.
For a discovery group, the ability to process more samples in less time should translate into reduced direct costs and faster results. Additionally, and importantly, generating more data faster can provide the time needed to carry out additional assays, providing deeper information to a discovery program on a group of compounds. The flexibility and capacity afforded by increased throughput also translates into the ability to spend more time working with problematic samples offline of the standard analytical process. These include potentially valuable samples that otherwise might be disregarded.
By turning the data around faster, the chemistry is faster, and presumably, the right drug can go into development faster. This undoubtedly will have a positive impact on productivity and efficiency in the drug discovery process.
MALDI QqQ also provides additional flexibility. By decoupling the chromatography and analytical steps, it simplifies the entire process — removing complexity, solvent, and consumable usage — and provides more options for timing experiments (that is, the biology in relation to the chemistry) and industrializing standard assays.
Less obviously, the use of MALDI plates provides a stable sample set. Also, because sample is not consumed completely in the first pass, the plate can be stored and re-run under different conditions. Adding the flexibility benefit, plates can be created in one location and analyzed in another.
In future applications, throughput increase will allow more time to develop better predictive assays. It could facilitate the move of safety–toxicology assays to MS, or it could allow for the development and application of improved in vitro assays that use more predictive markers for drug action–disease.
Other applications already being investigated include early discovery lead screening with enzyme assays and inhibitor screening (6), in which the high specificity of MS will enhance confidence in results greatly. Excitement is building about the potential applications of MALDI QqQ platforms for the future.
References
(1) J. Corr, P. Kovarik, B.B. Schneider, J. Hendrikse, A. Loboda, and T. Covey, J. Am. Soc. Mass Spectrom . 17, 1129–1141 (2006).
(2) L. Sleno and D.A. Volmer, Rapid Commun. Mass Spectrom . 19, 1928–1936 (2005).
(3) J. Gobey, M. Cole, J. Janiszewski, T. Covey, T. Chau, P. Kovarik, and J. Corr, Anal. Chem . 77, 5643–5654 (2005).
(4) Kent Beck and Cynthia Andres, Extreme Programming Explained — Embrance Change, Second Edition (Addison-Wesley Professional, Boston, 2004).
(5) Jakob Nielsen, Usability Engineering (Morgan Kaufmann, San Francisco, 1994).
(6) K.D. Greis, Mass Spectrom. Rev. 26, 324–339 (2007).
Jay Corr, Feng Zhong, Pauline Vollmerhaus, and Jean-François Alary are with MDS Analytical, Concord, Ontario. Daniel Lebre and Hesham Ghobarah are with Applied Biosystems, Concord, Ontario.

High pH and Temperature with HALO® ELEVATE C18, 1.5 mm ID Column
December 11th 2024Over the course of 5000 injections, the HALO®Elevate C18 1.5 mm ID column demonstrates stability and robustness in the stationary phase and the column hardware, with no significant changes to the retention time, peak shape, and back pressure.







