Analytical Characterization of Biotherapeutic Products, Part II: The Analytical Toolbox
LCGC North America
The analytical techniques used for characterizing biotherapeutics have evolved. We review the utility of the traditional tools and discuss the new, orthogonal techniques that are increasingly being used.
In the first part of this series, we discussed the various quality attributes that are pertinent to a biotherapeutic. In this issue, we will present the current and evolving practices that are being used for analysis of these attributes.
With increasing importance being attached to structural attributes of biotherapeutics and subsequent biosimilars, the analytical techniques used for characterizing these attributes have also evolved. There is an ever-increasing interest in attaining a higher structural resolution of these products (1,2). In view of the complexity exhibited by biopharmaceutical products, it has become the norm to use a multitude of orthogonal, high-resolution tools for characterization. These tools are carefully chosen such that the platform covers all the critical structural, physicochemical, immunochemical, and biologically relevant characteristics of the molecule (3–5). Attributes to be covered in biotherapeutic characterization, as per World Health Organization (WHO) guidelines to evaluate quality, safety and efficacy (2), are listed in Table I. This review focuses on the utility of the traditional characterization tools and covers the new generation of analytical hardware that is increasingly being used orthogonally to the traditional toolbox.
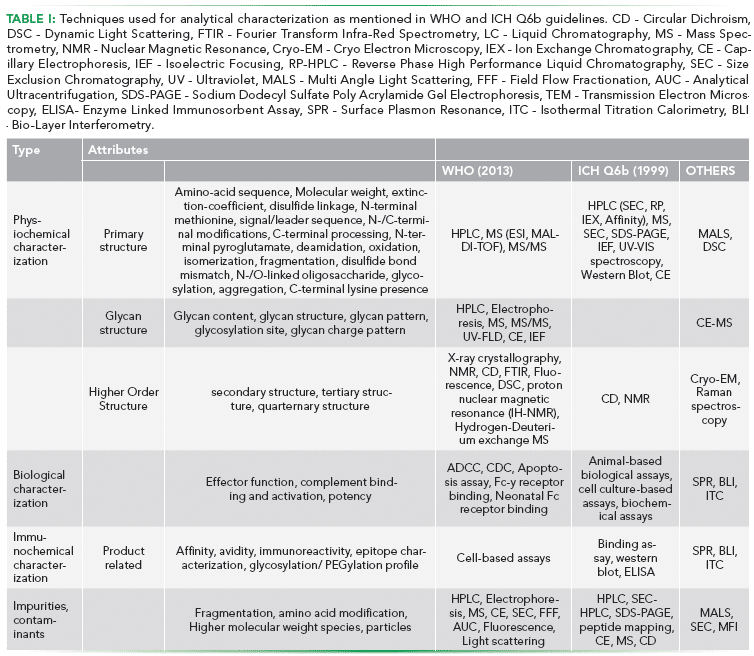
Tools for Analytical Characterization
Cost-effectiveness and relative ease of use have made Poly-Acrylamide Gel Electrophoresis (PAGE) one of the most commonly used techniques for protein analysis. In the case of biotherapeutic characterization, PAGE is typically used to estimate the size and isoelectric point of the molecule. It serves both as a detecting as well as resolving technique, and relies on the property of charged molecules to migrate in an electric field. Proteins are resolved on a polyacrylamide matrix, either in their native form (non-denaturing PAGE) or in reduced form (SDS-PAGE). A combination of the two can also be employed for resolving complex molecules such as monoclonal antibodies (mAbs), where separation from other proteins is attained in a non-denaturing first dimension followed by a denaturing second dimension resolution (2D-PAGE). Iso-Electric Focusing (IEF), which resolves molecules based on their iso-electric point is also a commonly used platform for 2D-PAGE and provides higher resolution, either in the first or the second dimension when compared with 2-D separation based only on size. Using Trastuzumab as an example, 2D-PAGE has also been shown to be a quick and easy method for qualitative evaluation of charge heterogeneity, stability and post-translational modifications. (6).
An interesting recent introduction in the 2D platform has been of 2D-Difference Gel Electrophoresis (2D- DIGE). Proteins are directly labelled with fluorescent dyes (CyDyes), pooled and separated on a 2D-PAGE. The dye binds covalently to ε-amino groups of lysine residues in proteins, allowing for accurate quantification of spots. The gels are scanned using an instrument capable of detecting different CyDye independently. 2D-DIGE can further be coupled with mass spectrometry for protein identification (7).
A major advantage of PAGE is its versatility. Gel matrix can be easily modified to achieve a specific resolution. Gradient gels (pH gradient) are routinely used to ascertain the isoelectric point. Moreover, PAGE can be coupled with Mass Spectrometry for identification on specific gel bands (first dimension) or spots (second dimension) (8, 9).
High resolution, varied choice of phase and robustness have made High Performace Liquid Chromatography (HPLC) a cornerstone of biopharmaceutical analysis. It is ubiquitously used for analysis of proteins, nucleic acids, or small molecules in complex mixtures. Typical separation is achieved by using a liquid mobile phase and a solid stationary phase. Diversity in solid phase chemistry and choice of mobile phase allows the analyst to fine-tune the type of interactions allowed between the analyte and the stationary phase. This yields unparalleled selectivity between a biotherapeutic and related variants and impurities, which otherwise may have near identical physicochemical properties. Some of the commonly used modalities of HPLC include:
- Ion Exchange (IEX) Chromatography is used to separate molecules based on their total charge. It enables the separation of molecules based on their charge. The strength of binding is determined by the affinity of the proteins to the "Ion-Exchanger" linked to the resin (stationary phase). Cationic exchangers possessing negative charge bind positively charged entities, whereas anionic exchangers bind negatively charged entities. Furthermore, ion-exchangers can be weak or strong depending upon the range of pH within which they can sustain their charge. This influences the range and type (strong/weak) of binding that can be achieved (10). Elution is typically achieved by either altering the pH of the mobile phase or increasing the ionic concentration of the mobile phase (salt gradient IEX). A commonly used application involves separation of charged variants, which are similar in size but differ in charge, using cation exchange HPLC (11).
- Size Exclusion Chromatography (SEC) enables separation of molecules based on their size. The stationary phase consists of a gel containing beads of a specific pore distribution. The choice of pore distribution allows the user to achieve separation of species in the desired range of size. In the case of biotherapeutics, a popular application is the resolution of size based heterogeneities, especially aggregates (12). Co-eluting host cell proteins can also be resolved using SEC (9).
- Hydrophobic Interaction Chromatography (HIC) separates molecules based on their hydrophobicity and their charge. It is a relatively gentle separation technique as the chosen conditions are minimally denaturing and as a result do not significantly affect the biological activity of the protein. Traditionally used as a polishing step in monoclonal antibody purification, it has also been used to characterize drug distribution in antibody-drug conjugates (ADCs) by exploiting the hydrophobicity of the conjugated small molecule (13).
- Reverse Phase (RP) Chromatography exploits reversible adsorption of biomolecules based on their hydrophobicity under conditions where the stationary phase is more hydrophobic than the mobile phase. It is quite similar to HIC in principle, however, the RPC medium is much more hydrophobic than in HIC and elution is achieved by the use of non-polar, organic solvents. This makes it a very desirable technique for coupling with MS for peptide mapping and other comparability studies. Coupled with mass spectrometry, the reverse phase has been extensively used in primary structure characterization of biotherapeutics as well as in comparability studies of biosimilars (14).
High Performance Capillary Electrophoresis (CE) is another technique increasingly being applied in conjunction with MS for charge variant characterization, isoelectric focusing and biosimilarity assessment. Molecules are separated across a fine capillary with the internal diameter as small as 50 µM via application of high voltage (~30 kV). This miniaturized format requires minimal sample with flow rates in the range of nL/min, making it a cost-effective technique. CE coupled with nano-ESI-MS has been shown to be particularly useful for N-Glycan analysis of monoclonal antibodies as well as for detection of charge variants (15). The availability of various CE modes such as capillary zone electrophoresis, capillary gel electrophoresis, capillary isoelectric focusing and micellar electrokinetic chromatography allows for characterization of different attributes such as intact mass, reduced mass, charge variants, as well as glycosylation pattern. CE coupled with MS is increasingly being used as a complementary platform to traditional LC-MS for biotherapeutic characterization (16). Innovative integration of CE with MS, such as in ZipChip has reduced the analysis time to under three minutes and bypassed issues of individual component integration and capillary damage due to handling. Demonstrated applications include glycosylation profiling of mAb directly from cell culture with sample volume requirement of under fifty microliters (17). The technique shows much promise in monitoring batch to batch variation in manufacturing as well as in biocomparability exercise.
X-ray crystallography is considered the gold standard for protein structural studies. It works on the principle of X-ray diffraction. The angle and the intensity of the diffracted beam from a crystal are used to construct a 3-dimensional image of the molecular structure. However, application of this technique for characterization and biocomparability is challenging as the protein of interest has to be purified and crystallized, which may not be possible for biotherapeutics due to the presence of inherent heterogeneity in the form of several post-translational modifications (PTMs). Although the data obtained is a direct measurement of the crystal structure, the technique itself is too cumbersome to be used as a routine analytical technique for biosimilar characterization and comparability (18).
Nuclear Magnetic Resonance (NMR) exploits the magnetic properties of specific atomic nuclei. Although a gold standard for protein structural studies, its routine application in the biotherapeutic industry has been limited due to several factors, including the large size of protein biopharmaceuticals, the relatively low sensitivity of the NMR signals, and the low natural abundance of active nuclei. However, in instances where biotherapeutics of small molecular size are being characterized, NMR might be utilized to gain in-depth HOS information (18).
It should be noted that although both X-ray crystallography and NMR provide structural details at a near-atomic level, the techniques differ in their principle, information and bottlenecks. X-ray crystallography requires the formation of the uniform crystal lattice and utilizes very high energy X-rays for deflection by the surrounding electron clouds, NMR is based on the absorption of electromagnetic radiation in the radio-frequency (RF) range. In NMR, proteins are analysed in solution and the final image is a compilation of a number of low energy states of the protein in different orientations as compared to single instance images of X-ray crystallography. Both the techniques require high concentration, homogenous sample preparations. With respect to protein size, NMR is more limiting. Proteins with a size larger than 40 kDa exhibit a slower molecular tumbling in solution leading to spectral overlap and peak broadening. In terms of information obtained, NMR can unravel information on structural dynamics and flexibility more readily than x-ray crystallography which requires time-resolved experiments to capture structural changes due to biochemical reactions (19, 20)
Transmission Electron Microscopy (TEM) is a microscopy technique in which a beam of electrons is transmitted through a specimen to form an image. Due to the much smaller wavelength of electrons as compared to light, the resolution of the image is greater by orders of magnitude with details up to atomic level, and hence TEM finds use in the characterization of biotherapeutic aggregates (21). Recent developments in achieving greater resolution, especially in Cryo-TEM, have enabled researchers to observe protein complexes such as mAbs in their native formulation (without crystallization) (22). The technique is currently underutilized in the field of biotherapeutics but has significant potential to grow.
Circular Dichroism (CD) Spectroscopy is based on the difference observed in the absorption of left and right-handed circularly polarized light of a molecule in the presence of light absorbing chiral groups. This property of biomolecules is frequently utilized in the examination of the secondary structure of the proteins and is employed for assessing HOS comparability. Using thermal denaturation, information about molecule stability as well as folding and unfolding mechanisms can be elucidated. An application of CD in the aggregate characterization of mAbs has also been demonstrated (23, 24).
Fourier Transform Infrared Spectroscopy (FTIR) is a spectroscopic technique that monitors the characteristic infrared absorption of molecules and translates it into structural information. Similar to CD, it gives structural information about the secondary structure of the protein, mainly the alpha helices and beta sheet. It serves as an orthogonal technique to CD in characterization and biocomparability studies (25).
Although both CD and FTIR can provide information with regards to components of the secondary structure of a molecule, it is only through higher resolution visualization techniques such as CryoTEM, X-ray crystallography and NMR that the specific arrangement of these components in space can be unravelled.
Dynamic Light Scattering (DLS) is commonly used to determine the size distribution profile of small particles in biotherapeutic formulations. The particle size profile is determined by measuring the random changes in the intensity of light scattered from the sample solution. It is often used in conjunction with SEC and analytical ultracentrifugation for aggregate studies. An interesting application of High-throughput platform in DLS (HT-DLS) has been in screening studies to quantify viscosity of mAb formulations (26). Using automated HT-DLS, effects of buffer conditions and temperature on aggregate formation have also been reported (27)
Raman Spectroscopy is used to observe vibrational, rotational, and other low-frequency modes in a system, generally applied to study the secondary structure of proteins by studying the Amide I band between 1600 and 1700 cm-1. An advantage that Raman spectroscopy offers over other secondary structure characterizing techniques is its low susceptibility to water interference as it detects scattered light and water is inefficient at scattering in this part of the spectrum (28). In a recent study, Raman Optical Activity (ROA) was evaluated as a means to detect early thermal instability in mAb samples kept at 50 °C for a month. Significant structural changes could be observed at one week of stress. This provides an advantage over SEC in monitoring aggregation as ROA would provide information about subtle differences in tertiary structure whereas Raman/ROA spectra can elucidate on changes at the secondary structure level (29). Another interesting recent application of Raman spectroscopy has been in drug identification, specifically in the identification of monoclonal antibodies by exploiting subtle differences in vibrational modes of the antibodies (30). It would be interesting to see if this application can be further modified to distinguish between biosimilars and innovator product.
Analytical Ultracentrifugation (AUC) is a versatile tool for quantitative analysis of macromolecules in solution. It employs the principle of centrifugal acceleration to separate particles based on their size and mass. Two types of hydrodynamic analyses, namely sedimentation velocity and sedimentation equilibrium, are able to make distinctions in formulation components based on shape and mass or mass alone, respectively. Sedimentation velocity is a mode of choice for aggregate analysis because it causes physical separation of molecular species of different mass or shape. This is a matrix-free technique as no column or gel matrix is required for size fractionation to occur (31). AUC-Sedimentation Velocity (AUC-SV) is often used to quantify high molecular weight species present in biopharmaceuticals (32).
Multiple Angle Light Scattering (MALS) is typically used in line with a resolving technique such as SEC to determine the size of the different molecular species as they pass through the MALS detector. It adds a quantitative measure of analysis to techniques like SEC. As the sample passes through the laser beam, light is scattered at multiple angles and the detector collects this data to approximate the sample size (33). It has been used to monitor the formation of soluble, HMWS so as to better quantify and model non-native aggregation kinetics in α chymotrypsinogen (34). Composition-Gradient Multi-Angle Light Scattering (CG-MALS), a variation on MALS, employs a series of unfractionated samples of different composition or concentration in order to characterize a wide range of macromolecular interactions. CG-MALS, a complementary technique to AUC and DLS, has been used to characterize self-association in model mAb molecule (35). Although not yet commonly used, CG-MALS could be used orthogonally to Surface Plasmon Resonance (SPR) to study receptor binding kinetics in biotherapeutics. Unlike SPR, CG-MALS would not require binding of the receptor to any chip and hence would be a truly label-free technique to quantify interactions, similar to Isothermal Titration Calorimetry (ITC). Although the latter is more suited to study the thermodynamic parameters of an interaction.
Field Flow Fractionation (FFF) is a unique separation technique used for analysis of aggregates. Samples are pumped into a narrow tube perpendicular to the flow and separation occurs due to the difference in mobility of the species in the mixture under the field applied. Depending upon the properties of the species in the sample mixture, different flows such as electrical, magnetic, thermal-gradient, gravitational or centrifugal can be applied. FFF serves as a complementary technique to DLS and MALS for determining the presence of sub-micron particles in the sample (36).
Fluorescence spectrometry is based on the principle that certain molecules called fluorophores emit light upon excitation by an external source such as an incandescent lamp or a laser which produces a spectrum. This technique is helpful in exploring HOS of a protein (to an extent), mainly the tertiary structure via the intrinsic fluorescence of the protein. Changes in the local environment of tryptophan, the strongest intrinsic fluorophore, are reflected in the emission spectra of the molecule and are a measure of change in the protein tertiary structure (37). In cases where a biomolecule does not have an intrinsic fluorophore, extrinsic fluorophore dyes such as Thioflavin-T (ThT) or 1-anilino-8-naphthale-nesulfonate (ANS) can be used to induce fluorescence. Fluorescence is routinely used in thermal stability studies of biotherapeutics, especially monoclonal antibodies (38). At best, it is an indirect measure of changes in a protein's tertiary structure and does not provide information about position and the specific nature of these changes.
Micro-Flow Imaging (MFI) is an up and coming technique that combines digital microscopy with microfluidics to capture and quantify sub-visible particles in the range of 1 to 300 µm in a solution. It can provide information about particle size, concentration and morphology (39). However, rather than protein characterization, it is more suited for profiling and classifying the particulate size in a given formulation.
Mass Spectrometry (MS) is a powerful and data-intensive technique for determining protein mass (intact, fragmented and reduced), sequence, and for probing and quantifying protein modifications. MS involves ionization of the sample fragments followed by their separation based on their mass to charge ratio (m/z). By accelerating the ionized particles and subjecting them to an electric or magnetic field, the ions get deflected depending on the mass and charge that they carry. Ions are detected by an electron multiplier and the results are displayed as a spectrum of the relative abundance of different ions based on the m/z ratio.
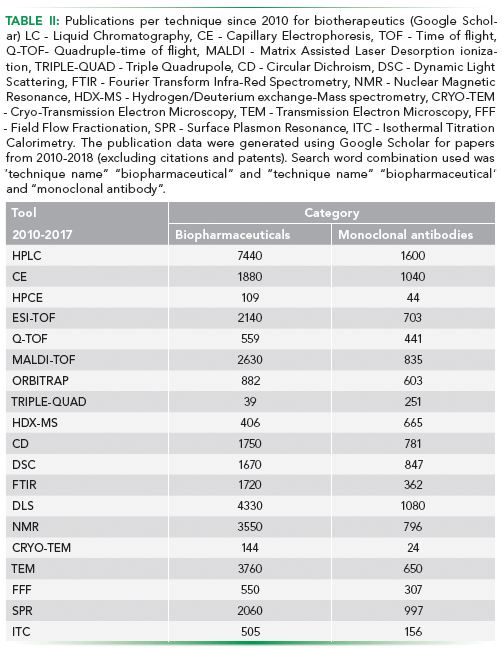
Analytical capability of an MS platform is defined by the kind of ionization source and the type of mass analyser being used. Examples of ionization sources include fast atom bombardment (FAB), chemical ionization (CI), atmospheric-pressure chemical ionization (APCI), electrospray ionization (ESI), and matrix-assisted laser desorption/ionization (MALDI). Examples of mass analyzers include Time-of-flight (TOF), quadrupole mass filter, and ion-traps. Some of the popular combinations of the two are ESI-TOF, MALDI-TOF and ESI-Q-TOF (Table II).
MS is usually coupled with LC or CE as the first dimension of separation. Characterization of complex molecules such as monoclonal antibodies requires mapping of the different fragmentation patterns in MS such as Electron Transfer Dissociation (ETD) and Collision-Induced Dissociation (CID) along with the use of different proteases make it possible to map disulphide links present and the different glycans attached to the protein (18,40).
Another useful application of MS, when coupled with Hydrogen/Deuterium exchange (HDX) is in elucidating protein conformational dynamics and protein interactions. In the biopharmaceutical industry, HDX-MS has established itself in the analysis of protein-small molecule interactions, characterization of bio-therapeutics/biosimilars, and epitope mapping of biotherapeutics. The technique relies on isotope labelling to probe the rate at which protein backbone amide hydrogens undergo exchange. MS is then used to monitor the mass-shift as a result of incorporation of deuterium in the protein. The rate of exchange provides information regarding the conformational mobility, hydrogen bonding strength, and solvent accessibility in protein structure (41,42)
Differential Scanning Calorimetry (DSC) is a versatile technique that measures the quantity of heat radiated or absorbed by the sample on the basis of a temperature difference between the sample and the reference material. It is used to determine equilibrium thermodynamic stability and folding mechanism of proteins and finds routine use in thermal stability characterization of biotherapeutics (43).
Tools for Functional Characterization:
Enzyme-linked Immunosorbent Assay (ELISA) is a popular diagnostic technique used to assess the immunogenicity of a therapeutic product. In a typical assay, a ligand, typically an antigen, is non-specifically or specifically bound to the polystyrene well of a 96 well microtiter plate. Enzyme-linked antibodies are used for colorimetric detection of a positive interaction upon addition of the enzyme substrate. ELISA has several applications in biotherapeutic characterization. A common application of this assay format is in Host Cell Protein analysis where polyclonal antibodies raised to the host cell are used to detect the presence of HCPs in the sample (44). However, with high specificity techniques such as MS (detection) and Surface plasmon resonance (interaction) becoming affordable and routine, ELISA can only be used for a precursor technique for quick estimation with limited confidence prior to employing tools with much higher specificity.
Surface Plasmon Resonance (SPR) (interaction) is a label-free technique that allows for real-time detection of biomolecular interactions. The SPR phenomenon occurs when polarized light strikes an electrically conducting surface at the interface between two media. In response, plasmons are produced, which are electron charged density waves. These waves reduce the intensity of reflected light at a specific angle known as the resonance angle, in proportion to the mass on a sensor surface. SPR allows for real-time monitoring of both association and dissociation of an interaction, generating reproducible kinetic data. Due to its sensitivity, ease of use and automated data analysis, it is widely used to study the ligand-receptor kinetics of monoclonal antibodies (14)
Bio-Layer Interferometry (BLI) is a label-free, optical analytical technique that analyzes the interference pattern of white light reflected from two surfaces: a layer of immobilized protein on the biosensor tip and an internal reference layer. Changes occurring in the number of molecules bound to the biosensor tip is reflected as a shift in the interference pattern. Similar to SPR, binding kinetics can be determined by this technique. Because of its robustness and ease of implementation, BLI is gaining application as a complementary technique to SPR for studying and comparing biotherapeutic binding kinetics (44).
Isothermal Titration Calorimetry (ITC) is a physical technique used to determine the thermodynamic parameters of interactions in a solution. It measures the heat released or absorbed by mixing the two interactants via titration. It is widely considered an absolute and direct measurement of interaction. Thermodynamic parameters of an interaction can be assessed via this technique and it is fast gaining importance as an orthogonal technique to SPR for ligand binding studies in biocomparability (45).
Summary
With the increase in complexity of the biotherapeutics undergoing manufacturing, the need for in-depth analytical characterization has also increased. As protein molecules, even minute changes in the structure of the biotherapeutic can confer altered functionality, sometimes leading to immunogenic reactions in the patients. In view of our dependency for production of these molecules on biological machinery, the formation of numerous altered conformations of the molecule is unavoidable. However, significant advancements have been made in analytical methodology increasing our ability to characterize a biotherapeutic. This is highlighted by the increase in the number of technique rich publications on analysis of biotherapeutics since 2010 and the introduction of the US-FDA BPCI Act 2009 (Table II).
Our understanding of the relevance of the different measured attributes with respect to the safety and efficacy of a biotherapeutic has been improving with time and experience. Some recently evolving techniques such as CG-MALS for macromolecular interactions and intrinsic Förster resonance energy transfer (iFRET) for in vivo target protein detection are yet to find their place in the biotherapeutic characterization toolbox while other well-established techniques such as Raman spectroscopy are being increasingly used for novel applications such as drug identification. A desirable direction for technological advancements would be the increasing throughput of techniques such as LC by parallelization (9). Short analysis time and micro- and nanoplatforms with minimal sample consumption would help cut down the cost of analysis associated with limited and expensive samples such as monoclonal antibodies.
Despite these advancements, we are yet to reach a point where a product such as a complex biotherapeutic, can be completely fingerprinted in a manner similar to a pharmaceutical (small molecule) product. Moreover, each technique comes with its unique limitations and pitfalls. This ensures that the topic of analytical characterization of biotherapeutics will continue to be an area of research in the time to come.
References
(1) ICH Expert Working Group, Specif. Test Proced. Accept. Critreia Biotechnol. Prod. (March), 1–20 (1999).
(2) WHO, Guidelines on the Quality, Safety and Efficacy of Biotherapeutic Protein Products Prepared by Recombinant DNA Technology, WHO Technical Report Series 814, 91 (2013)
(3) L.A. Bui, S. Hurst, G.L. Finch, B. Ingram, I.A. Jacobs, C.F. Kirchhoff, C.K. Ng, and A.M. Ryan, Drug Discov. Today20(S1), 3–15 (2015).
(4) A.S. Rathore, Trends Biotechnol. 27(12), 698–705 (2009).
(5) A. AL-Sabbagh, E. Olech, J.E. McClellan, and C.F. Kirchhoff, Semin. Arthritis Rheum. 45(5), S11–S18 (2016).
(6) D. Nebija, C. Noe, E. Urban, and B. Lachmann, Int. J. Mol. Sci. 15(12), 6399–6411 (2014).
(7) R. Diez, M. Herbstreith, C. Osorio, and O. Alzate, 2-D Fluorescence Difference Gel Electrophoresis (DIGE) in Neuroproteomics Neuroproteomics (CRC Press/Taylor & Francis, 2010). at <http://www.ncbi.nlm.nih.gov/pubmed/21882449>.
(8) C. Reichel and M. Thevis, Bioanalysis 5(5), 587–602 (2013).
(9) S. Fekete, D. Guillarme, P. Sandra, and K. Sandra, Anal. Chem . 88(1), 480–507 (2016).
(10) J. R. Auclair, A. S. Rathore Chitra, and I. S. Krull, LCGC North Am. 36(1), 26–36 (2018).
(11) S.K. Singh, G. Narula, and A.S. Rathore, Electrophoresis 37(17–18), 2338–2346 (2016).
(12) A. Singla, R. Bansal, V. Joshi, and A.S. Rathore, AAPS J. 18(3), 689–702 (2016).
(13) A. Wakankar, Y. Chen, Y. Gokarn, and F.S. Jacobson, MAbs 3(2), 164–175 (2011).
(14) N. Nupur, N. Chhabra, R. Dash, and A.S. Rathore, MAbs 10(1), 143–158 (2018).
(15) K. Wooding, W. Peng, and Y. Mechref, Curr. Pharm. Biotechnol. 17(9), 788–801 (2016).
(16) M. Han, B. M. Rock, J. T. Pearson, Y. Wang, and D. A. Rock, Therapeutic Monoclonal Antibody Intact Mass Analysis by Capillary Electrophoresis–Mass Spectrometryin Capill. Electrophor. Spectrom. , 13–34 (Springer International Publishing, Cham, 2016). doi:10.1007/978-3-319-46240-0_3.
(17) S.A. Berkowitz, J.R. Engen, J.R. Mazzeo, and G.B. Jones, Nat. Rev. Drug Discov. 11(7), 527–540 (2012).
(18) S.A. Berkowitz, Analytical Characterizationin Biosimilar Drug Prod. Dev. , 15–82 (2017). doi:10.1201/9781315119878-3.
(19) B. Carragher, A. Schneemann, J.J. Sung, S.K. Mulligan, J.A. Speir, K. On, and C.S. Potter, Microsc. Microanal. 21(33), 2014–2015 (2015).
(20) J. T. Yang, C.-S. C. Wu, and H. M. Martinez, Methods Enzymol. 130, 208–269 (1986). doi:10.1016/0076-6879(86)30013-2.
(21) V. Joshi, T. Shivach, N. Yadav, and A.S. Rathore, Anal. Chem. 86(23), 11606–11613 (2014).
(22) L.R. Tsuruta, M. Lopes dos Santos, and A.M. Moro, Biotechnol. Prog . 31(5), 1139–1149 (2015).
(23) F. He, G. W. Becker, J. R. Litowski, O. L. Narhi, D. N. Brems, V. I. Razinkov, Anal. Biochem. 399(1), 141–143 (2010).
(24) L. Aileen, A. Seneviratne, G. Ratnaswamy, and J. Park, Pharm. Technol. 38(10), 32–39 (2015).
(25) R.J. Falconer, D. Jackson-Matthews, and S.M. Mahler, J. Chem. Technol. Biotechnol. 86(7), 915–922 (2011).
(26) G. Thiagarajan, E. Widjaja, J.H. Heo, J.K. Cheung, B. Wabuyele, X. Mou, and M. Shameem, J. Raman Spectrosc. 46(6), 531–536 (2015).
(27) S.K. Paidi, S. Soumik, R. Strouse, J.B. McGivney, C. Larkin, I. Barman, Anal. Chem. 88(8), 4361–4368 (2016).
(28) T. Arakawa, J.S. Philo, D. Ejima, K. Tsumoto, and F. Arisaka, Bioprocess Int. 4, 42–43 (2006).
(29) L. Wafer, M. Kloczewiak, and Y. Luo, AAPS J. 18(4), 849–860 (2016).
(30) A. Oliva, M. Llabrés, and J.B. Fariña, J. Pharm. Biomed. Anal. 25(5–6), 833–841 (2001).
(31) Y. Li, W. F. Weiss, and C. J. Roberts, J. Pharm. Sci. 98(11), 3997–4016 (2009).
(32) R. Esfandiary et al., J. Pharm. Sci. 102(9), 3089–3099 (2013).
(33) J. Liu, T. Eris, C. Li, S. Cao, and S. Kuhns, BioDrugs 30(4), 321–38 (2016).
(34) P. Garidel, M. Hegyi, S. Bassarab, and M. Weichel, Biotechnol. J. 3(9–10), 1201–1211 (2008).
(35) M. Weichel, S. Bassarab, and P. Garidel, Bioprocess Int. (5), 42–52 (2008). at <http://www.bioprocessintl.com/manufacturing/monoclonal-antibodies/probing-thermal-stability-of-mabs-by-intrinsic-tryptophan-fluorescence-182648/>.
(36) M.K. Joubert, Q. Luo, Y. Nashed-Samuel, J. Wypych, and L.O. Narhi, J. Biol. Chem. 286(28), 25118–33 (2011).
(37) A. Guttman, LCGC North Am. 30(5), 412–421 (2012).
(38) C.M. Johnson, Arch. Biochem. Biophys. 531(1–2), 100–109 (2013).
(39) D.G. Bracewell, R. Francis, and C.M. Smales, Biotechnol. Bioeng. 112(9), 1727–1737 (2015).
(40) U. Sinha-Datta, S. Khan, and D. Wadgaonkar, Biosimilars 5, 83–91 (2015).
(41) C.A. Challener, BioPharm Int. 28(1) (2015).
(42) T.K. Dam, M. Torres, C.F. Brewer, and A. Casadevall, J. Biol. Chem. 283(46), 31366–70 (2008).
(43) M.G. Petroff, H. Bao, J.P. Welsh, M. van Beuningen-de Vaan, J.M. Pollard, J.D. Roush, S. Kandula, P. Machielsen, N. Tugcu, and T. O. Linden, Biotechnol. Bioeng. 113(6), 1273–1283 (2016).
(44) V. Kamat and A. Rafique, Anal Biochem. 536, 16-31 (2017)
(45) S. Perspicace, A.C. Rufer, R. Thoma, F. Muller, M. Hennig, S. Ceccarelli, T. Schulz-Gasch and J. Seelig, FEBS Open Bio. 3, 204-211 (2013)
ABOUT THE AUTHORS
Anurag S. Rathore

Anurag S. Rathore is a professor in the Department of Chemical Engineering at the Indian Institute of Technology in Delhi, India.
Ira S. Krull

Ira S. Krull is a Professor Emeritus with the Department of Chemistry and Chemical Biology at Northeastern University in Boston, Massachusetts, and a member of LCGC 's editorial advisory board.
Srishti Joshi

Srishti Joshi is a post-doctoral research fellow in the Bioseparations and Bioprocessing Lab, under the tutelage of Professor Anurag. S. Rathore, Department of Chemical Engineering, Indian Institute of Technology, Delhi.













