Amino Acid Isomer Separation Using the Intrada Amino Acid Column from Imtakt
The Application Notebook
The Intrada Amino Acid column has been shown to be a viable solution for the analysis of amino acids by LC–MS without derivatization.
The Intrada Amino Acid column has been shown to be a viable solution for the analysis of amino acids by LC–MS without derivatization. This current work addresses the limitation of this detection method, where isomers and isobaric compounds may not have distinguishable m/z ratios.
Separation of amino acids isomers by LC–MS faces multiple challenges. First, these compounds may be isobaric (same m/z ratio), constraining the application of a mass spectrometer (MS). Secondly, they may have very similar molecular structure making them difficult to resolve chromatographically using current HPLC column chemistries.
Here we show that the unique stationary phase of the Intrada Amino Acid column can address these limitations, providing an excellent solution for the analysis of amino acid isomers by LC–MS. Specifically, this note presents data for leucine and threonine isomers.
Experimental Conditions
A 5 μL sample at 1 mM concentration in 0.1 N HCl was injected onto Intrada Amino Acid column. Eluent was analyzed using single quadrupole MS in ESI, positive mode. Further experimental details are provided as inserts in respective chromatograms.
Results and Discussion
Despite the fact that leucine isomers, Figure 1, are structurally similar, the Intrada Amino Acid column separates these compounds well. The difficulty with this separation is that they differ primarily only in the alkyl portion of their structure, specifically, the methyl group geometry. This challenge is exacerbated by the fact that the Intrada Amino Acid column has a normal phase mixed-mode stationary phase, which conceptually should struggle with separating analytes based solely on their hydrophobic portions. Despite this, the Intrada Amino Acid column is able to separate these isomers, further highlighting its extraordinary resolving power.
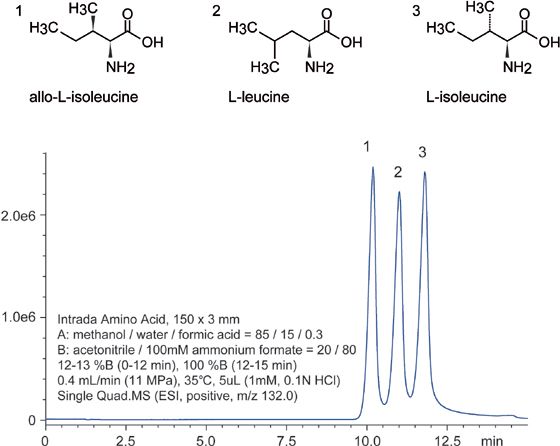
Figure 1: LC–MS separations of leucine isomers.
Threonine isomers separation is shown in Figure 2. Overall, the peaks are well-resolved (elution times of 11, 15, 16, and 18 min), with only a minimal coelution for peaks 2 and 3 observed. This separation provides an additional example of the resolving power the Intrada Amino Acid column in that the only difference between L-threonine and allo-L-threonine is in their three-dimensional shape, indicating the presence of steric selectivity in addition to the other modes indicated.
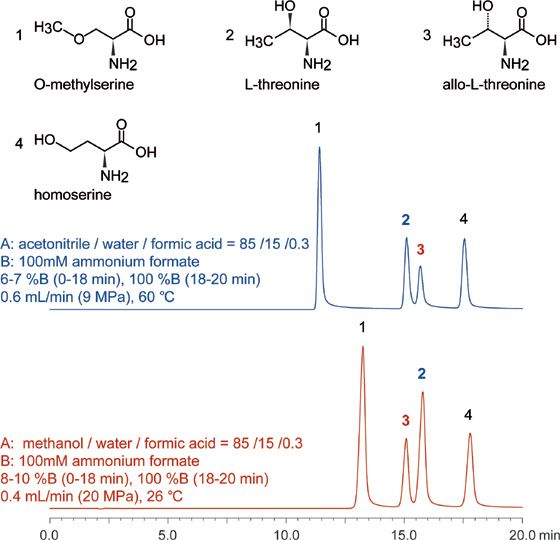
Figure 2: LC–MS separation of threonine isomers. Intrada Amino Acid, 250 × 3 mm.
Conclusion
In summary, the Intrada Amino Acid column has been shown to provide excellent separation power for underivatized amino acid isomers. Resolved compounds shown here include ones with distinctive m/z ratios, isomers containing minimal aliphatic differences, as well as only steric differences. These results are of particular importance as isomers are extremely challenging to separate using more conventional approaches, yet, the Intrada Amino Acid column shows adequate chromatographic resolution of both positional isomers and stereoisomers. Therefore, this evidence validates the use of this column for the separation of a wide range of amino acids when used in concert with mass spectrometry detection.

Imtakt USA
1104 NW Overton St., Portland, OR 97209
tel. (888) 456-HPLC, (215) 665-8902, fax (501)646-3497
Website: www.imtaktusa.com

SEC-MALS of Antibody Therapeutics—A Robust Method for In-Depth Sample Characterization
June 1st 2022Monoclonal antibodies (mAbs) are effective therapeutics for cancers, auto-immune diseases, viral infections, and other diseases. Recent developments in antibody therapeutics aim to add more specific binding regions (bi- and multi-specificity) to increase their effectiveness and/or to downsize the molecule to the specific binding regions (for example, scFv or Fab fragment) to achieve better penetration of the tissue. As the molecule gets more complex, the possible high and low molecular weight (H/LMW) impurities become more complex, too. In order to accurately analyze the various species, more advanced detection than ultraviolet (UV) is required to characterize a mAb sample.












