A Study on Optimum Method Conditions for Denaturated and Non-Denaturated siRNA by Ion Pair Reversed-Phase Liquid Chromatography
Small interfering ribonucleic acids (siRNAs) are a promising therapeutic approach, and ion pair (IP) reversed-phase chromatography is the gold standard used to analyze them. As siRNAs usually occur as duplexes, they can be analyzed under non-denaturing or denaturing conditions. In the latter case, both the sense and antisense strands can be monitored. Several parameters influence an siRNA separation. This article discusses the most important aspects: the column temperature, the type of IP reagent, and the corresponding acid. By using a bioinert column, the influence of the column hardware can be eliminated or at least reduced to a minimum.
Oligonucleotides have grown in importance for medical applications over recent years. The COVID-19 pandemic placed a focus on messenger ribonucleic acid (mRNA)-based vaccines. Other therapeutic oligonucleotides are based on antisense oligonucleotides (ASOs), micro-RNAs (miRNAs), and small interfering RNAs (siRNAs), which are used to trigger gene inhibition (1,2). siRNA is an important part of a naturally occurring mechanism to defend the invasion of foreign nucleic acids, called the RNA interference (RNAi), which was discovered in 1998 (2,3). Therefore, siRNAs are a promising therapeutic approach because they theoretically can silence any disease‑related genes in a sequence-specific manner. After 20 years of research the first siRNA drug, Patisiran (Onpattro, Alnylam)—used to treat a rare disease of polyneuropathy— was approved in 2018. To date, at least five different siRNA-based pharmaceuticals are approved by the US Food and Drug Administration (FDA) and/or the European Commission (EC) (3).
Robust and highly sensitive analytical methods are therefore required. siRNAs usually occur as oligonucleotide duplexes. They can be analyzed under non‑denaturing conditions so that duplex formation is assured and excessive single‑strands are detected. Alternatively, analysis of siRNAs can be conducted under denaturing conditions so that both sense and antisense strands are monitored. In addition to ion pair (IP) reversed-phase liquid chromatography (LC), which remains the gold standard for the characterization of oligonucleotides and their by-products, anion exchange chromatography (AEC) is an alternative approach (4,5). Furthermore, size‑exclusion chromatography (SEC) can be used to separate double‑stranded from single‑stranded siRNA (6).
Due to the electron-rich backbone of oligonucleotides, bioinert column hardware can be used for the analysis of siRNAs to suppress undesired ionic interactions (5). In order to achieve optimum results, a number of parameters have to be considered. Together with the type of ion‑pairing agent and the corresponding acid, column temperature is a key factor for the denaturation of siRNAs.
This article will provide an overview of important aspects for the analysis of siRNAs, with a special focus on non-denaturating relative to denaturating conditions.
Experimental Conditions
An siRNA (21mer) with the following sequence:
5’-CGU ACG CGG AAU ACU UCG AdTdT-3’ sense strand
3’-dTdTGCA UGC GCC UUA UGA AGC U-5’ antisense strand
as well as the corresponding two single‑strands as reference was used in this study. The general chromatographic conditions were kept constant as follows: flow rate 0.42 mL/min, UV detection at 260 nm, and injection volume 1 μL. The general gradient was 1% B/min, starting from 3% B unless stated otherwise. A 50 × 2.1 mm, 1.9‑μm widepore YMC‑Accura Triart Bio C18 UHPLC column with a bioinert coating was used. The eluents used were A) 8 mM triethylamine (TEA) containing 100 mM 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) if not stated otherwise, and B) methanol. Different temperatures of 65 °C, 45 °C, or 25 °C were studied.
The Effect of Column Temperature
The denaturation of oligonucleotides depends on various factors such as the temperature and modifiers used. The thermal denaturation of double‑stranded oligonucleotides, which is also called the melting point, usually proceeds at high temperatures (7). The denaturation temperature of siRNAs depends on their length and composition. On-column denaturation can be observed by a peak shift since oligonucleotide duplexes adhere strongly to the stationary phase so that single-strands elute earlier (8). Therefore, it is useful to determine the denaturation temperature of the RNA sample.
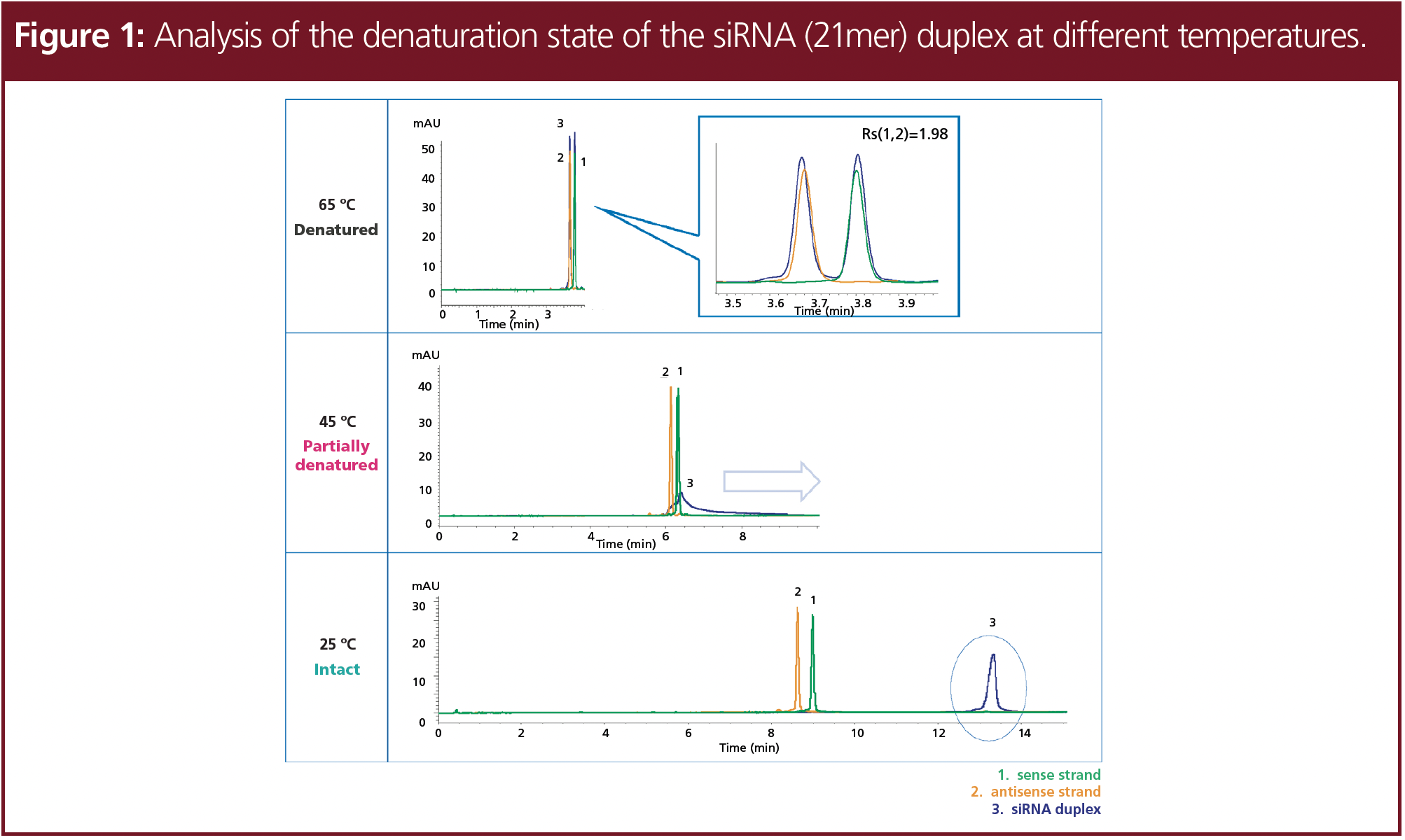
In Figure 1, the different states of the siRNA at different temperatures are demonstrated. At an elevated temperature of 65 °C two signals were detected, which correspond to the two single-strands, indicating that the siRNA duplex was fully denaturated into two single-strands. At 45 °C, the siRNA was partially degraded, which is shown due to peak distortion and a slightly increased retention. At 25 °C, non‑denatured siRNA was detected, which eluted significantly later compared to the single strands.
Type and Concentration of Ion-Pairing Agent
As a result of the electron-rich phosphate backbone of siRNA, the retention on common hydrophobic stationary phases is too low. Ion‑pairing agents such as n-alkylamines can help to overcome this challenge. The resulting ion pairs of n-alkylamine and oligonucleotide can be separated with a reversed-phase column according to their hydrophobicity (4).
n-Butylamine (BA), triethylamine (TEA), hexylamine (HA), and dibutylamine (DBA) were screened at three different temperatures corresponding to the temperature screening to find the ideal ion-pairing agent (see Figure 2). Based on previous studies, all ion-pairing agents were tested at a concentration of 8 mM in combination with 100 mM HFIP, resulting in a pH of around 8 (9). The initial eluent concentration of the gradient was adjusted individually to achieve comparable retention.
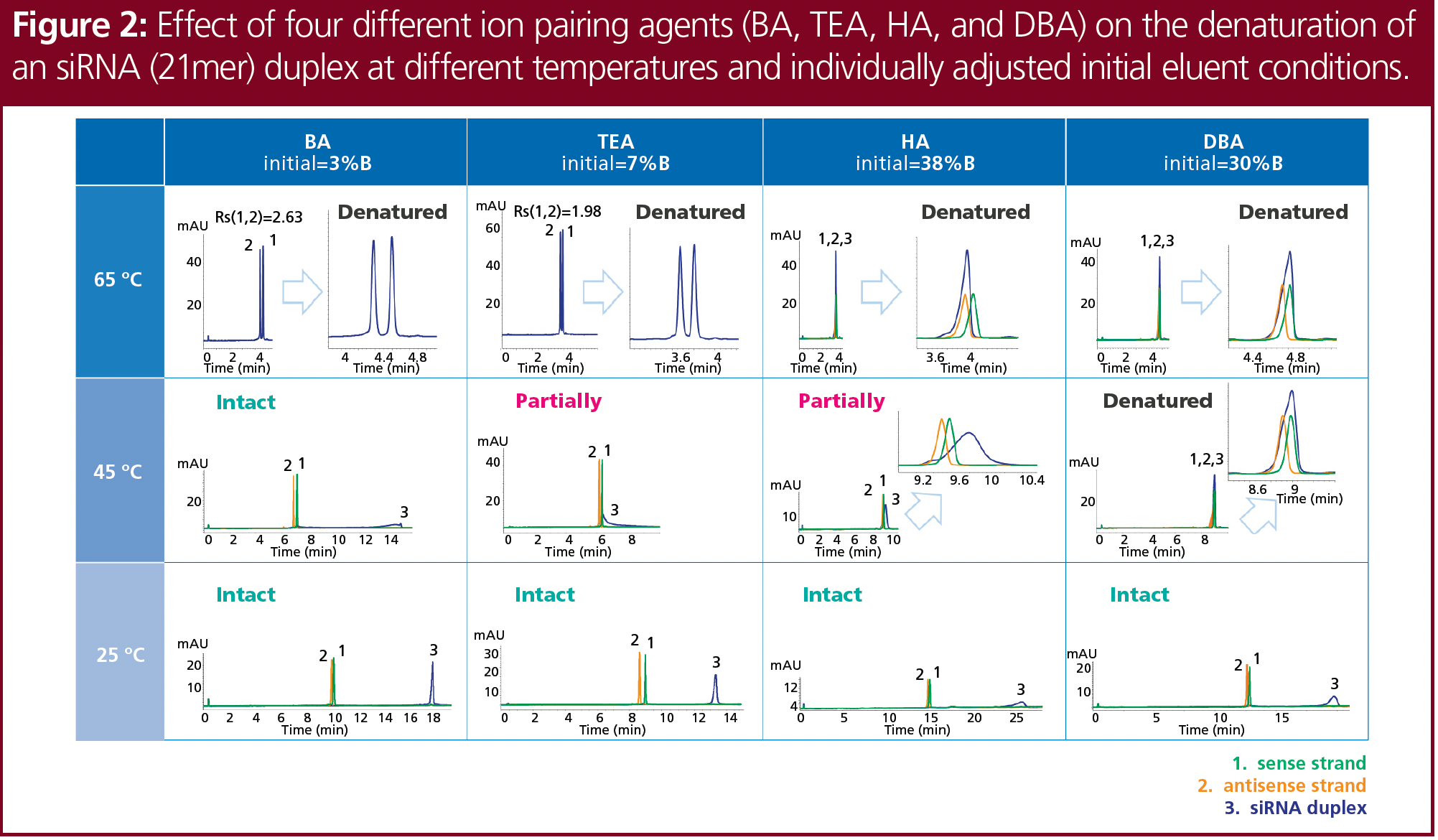
At 65 °C, the siRNA was denatured with all ion-pairing agents tested, but only BA and TEA provided an adequate separation of the two single strands; there was no separation obtained using HA or DBA. At 25 °C, no denaturation was observed and sufficient recovery and optimum peak shape were achieved using BA or TEA. At 45 °C, the siRNA partially denatured with TEA and HA, and completely denatured with DBA. No denaturation was observed with BA. Further research was performed using TEA, as it provided the best overall results—which included all three states of denaturation.
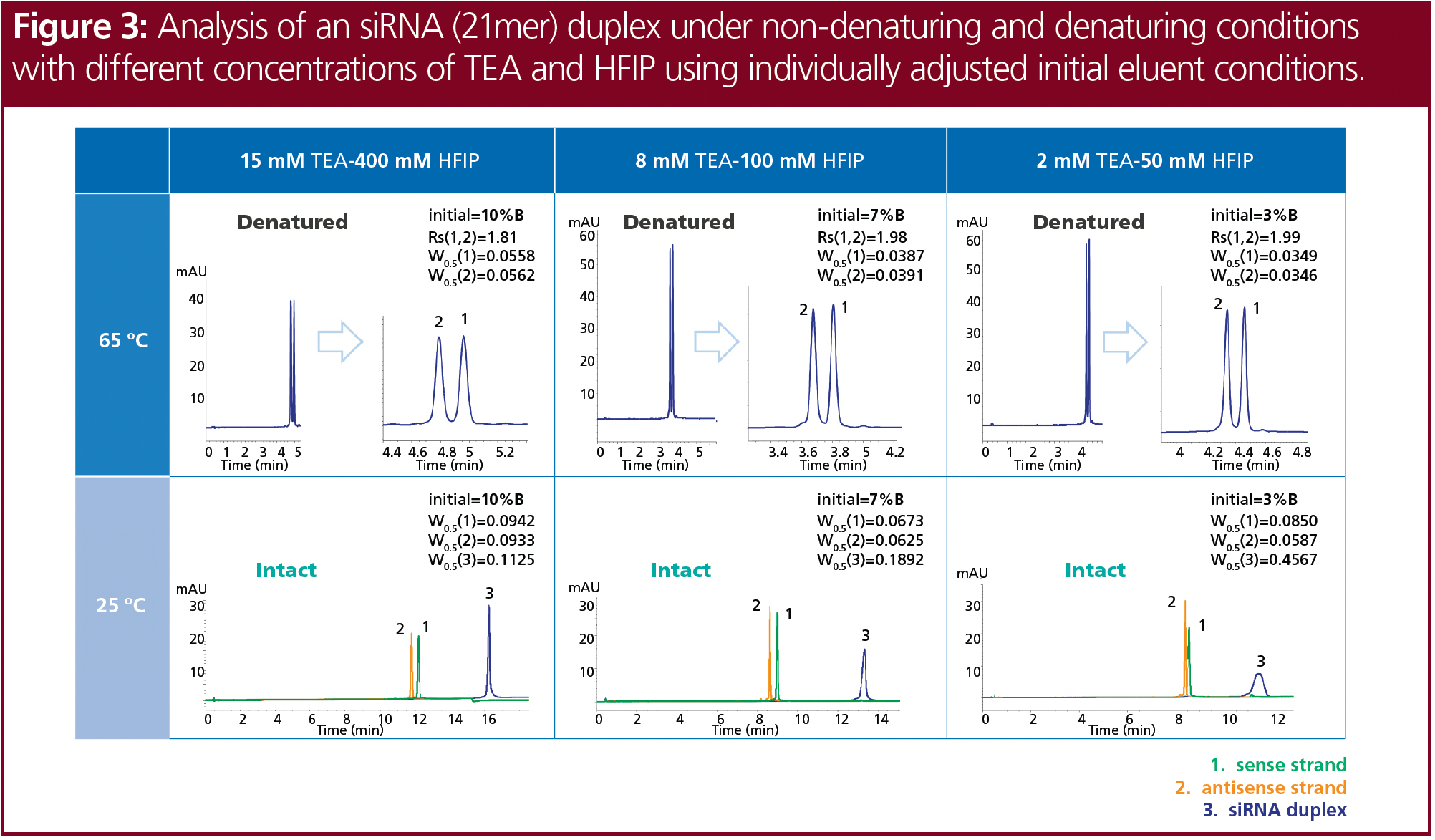
Mass spectrometry (MS) applications specifically require volatile ion-pairing reagents. Therefore, TEA in combination with HFIP as the acidic counter ion and methanol as organic modifier is a common choice (10). Different concentrations of TEA and HFIP at pH 8 were tested for the denaturated siRNA at 65 °C and the intact siRNA duplex at 25 °C (Figure 3). Again, the initial eluent concentration of the gradient was adjusted individually. Lower concentrations of TEA and HFIP slightly improved the resolution of the separated denatured siRNA. Better sensitivity and sharper peaks were obtained at low ion-pairing agent concentrations for the denaturated single strands, but the siRNA duplex showed significantly improved peak shape at higher concentrations of TEA and HFIP. In contrast to the applied UV detection, the lower concentrations may be better suited for use with MS detection.
Acetic Acid as Alternative for HFIP
In addition to TEA in combination with HFIP, TEAA buffer (TEA and acetic acid) is commonly used as an ion-pairing reagent. When high-throughput analyses are performed, costs can be reduced by exchanging HFIP with another counter ion. The TEAA buffer is also compatible with MS detection. The influence of acetic acid on oligonucleotide separation and denaturation was tested by comparing both ion-pairing agents (Figure 4). Two distinct eluents of 15 mM TEA with 400 mM HFIP and 15 mM TEAA buffer (pH 8) were compared. For the TEAA buffer, 15 mM TEA was adjusted with acetic acid to pH 8. The initial amount of eluent B was 7% for TEA–HFIP and 5% for TEAA.
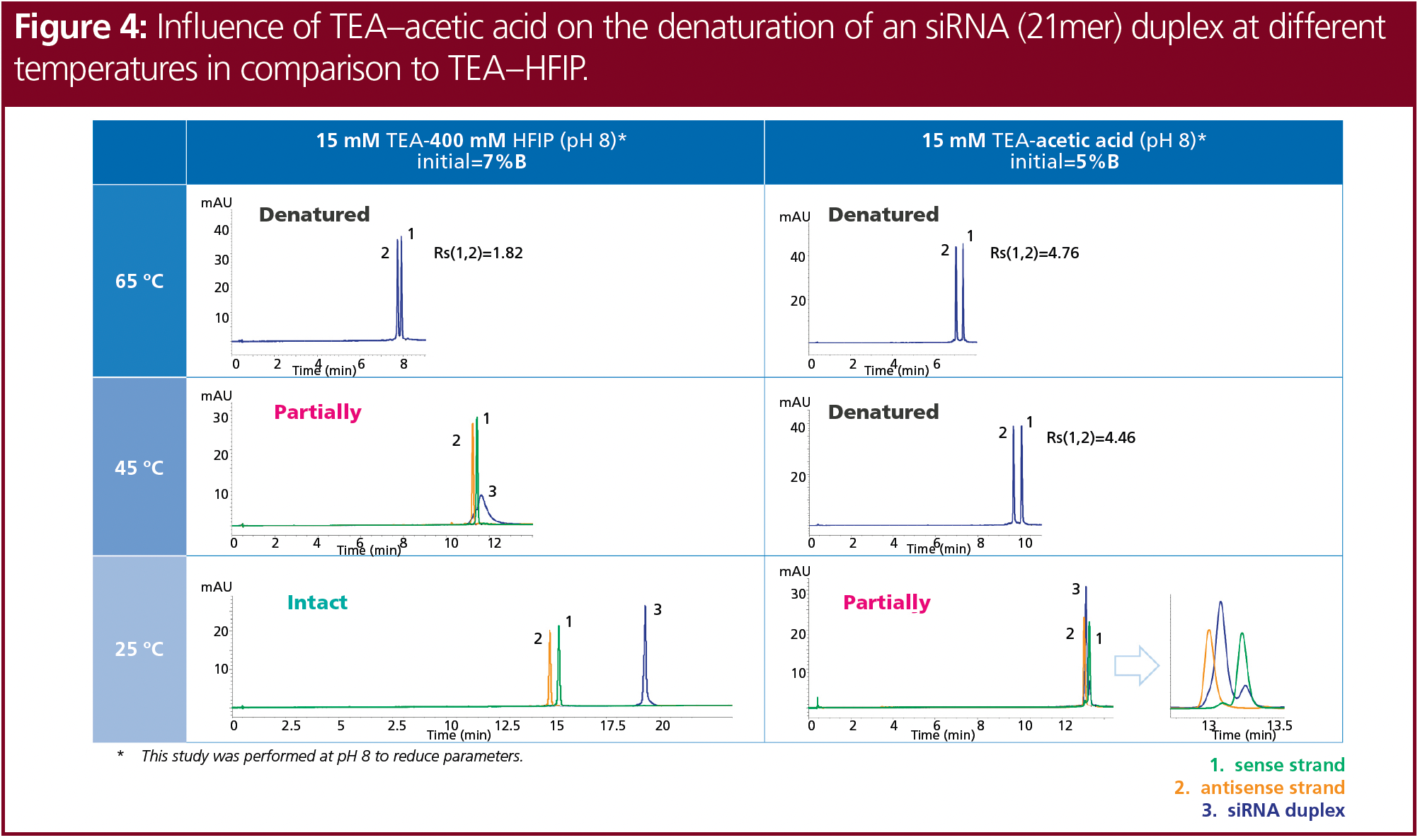
The use of TEAA resulted in better resolution of the denaturated single strands and slightly higher sensitivity at 65 °C compared to TEA–HFIP. At 45 °C, siRNA was fully denatured using TEAA but only partially denatured with TEA–HFIP. Moreover, at 25 °C the siRNA was only partially denatured when TEAA was used, whereas it was intact at these conditions when tested with TEA–HFIP. TEAA is therefore not a suitable alternative for TEA–HFIP for the analysis of intact siRNA duplexes. However, the temperature could be decreased to 45 °C to analyze the denaturated single strands with TEAA.
BA, HA, and DBA were tested as alternatives to TEA at 65 °C (not shown). In combination with HFIP as well as acetic acid, the best resolution was still seen using TEA. Combinations of HA and DBA with HFIP or acetic acid did not lead to a separation of siRNA or the respective single strands at all.
Optimum Conditions
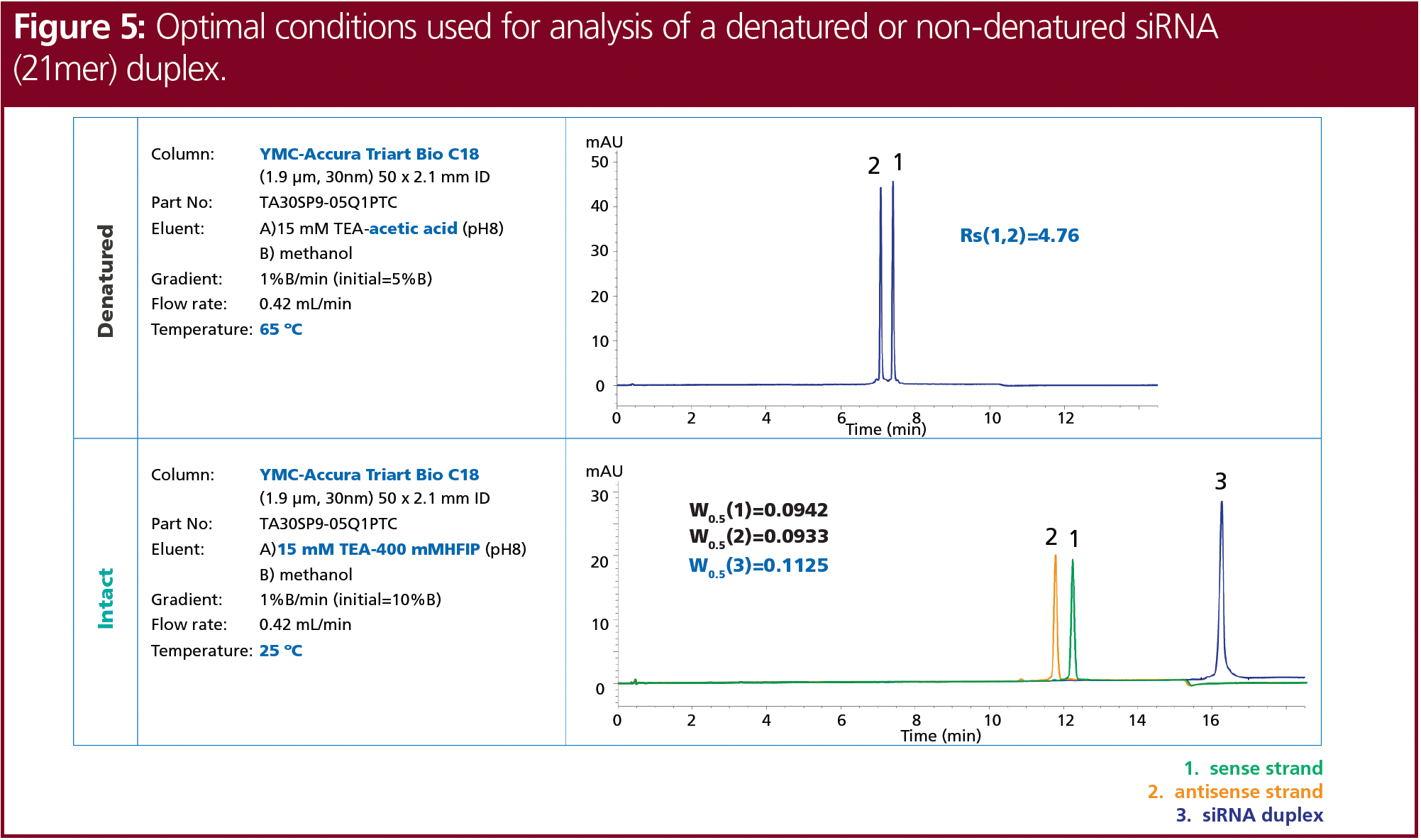
The optimum conditions for the denatured and non-denatured analysis of the siRNA tested were determined. The complete chromatographic conditions are shown in Figure 5. The siRNA was fully denatured and separated using 15 mM TEA adjusted to pH 8, with acetic acid and methanol as organic modifier at 65 °C. In contrast, the analysis of the intact siRNA duplex showed the best results using 15 mM TEA and 400 mM HFIP, with methanol at 25 °C. Nevertheless, the analysis of siRNA by ion‑pair LC must always be adjusted individually to the properties of the respective siRNA. Furthermore, the analyses were performed using UV detection. Detection using MS might need further adjustments.
Conclusion
In order to achieve optimum results for the analysis of siRNAs using ion-pair LC, a number of factors must be considered, including the column temperature, the selection and concentration of the ion-pairing agent, as well as the influence of the acidic counter ion. Furthermore, a bioinert column hardware and system should be used to suppress ionic interactions.
Ion-pair LC can be applied with MS detection, although ion-pairing agents have a strong influence on ionization efficiency and sensitivity. This limits the choice of suitable eluent modifiers. HFIP is well suited and its compatibility with MS has great advantages, but the higher cost is a disadvantage. Therefore, alternative counter ions, particularly for high-throughput analyses, need to be considered. Counter ions such as acetic acid can increase the denaturation of siRNAs. Nevertheless, the analysis of siRNA by ion-pair LC must always be individually adjusted to the properties of the respective siRNA. In this particular case, good resolution and retention times were obtained when the ion-pairing effect was enhanced, therefore applying high TEA concentrations at a pH of about 8 should be used. Column temperature is a crucial key factor to analyze siRNAs in a denaturated relative to a non-denaturated state. Therefore, an elevated temperature of 65 °C provided good results and full denaturation. The best resolution for the denaturated siRNA single strands was obtained when using a 15 mM TEAA buffer at pH 8. In contrast, for the analysis of the intact siRNA duplex, a low temperature of 25 °C and the use of 15 mM TEA and 400 mM HFIP provided the best peak shape and sensitivity.
References
(1) Hu, B.; Zhong, L.; Weng, Y.; et al. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 2020, 5, 101. DOI: 10.1038/s41392-020-0207
(2) Chernikov, I.V.; Vlassov, V.V.; Chernolovskaya, E.L. Current Development of siRNA Bioconjugates: From Research to the Clinic. Front. Pharmacol. 2019, 10, 444. DOI: 10.3389/fphar.2019.00444
(3) Friedrich, M.; Aigner, A. Therapeutic siRNA: State‑of‑the‑Art and Future Perspectives. BioDrugs 2022, 36, 549–571. DOI: 10.1007/s40259-022- 00549-3
(4) Gilar, M.; Fountain, K.J.; Budman, Y.; et al. Ion‑pair reversed-phase high-performance liquid chromatography analysis of oligonucleotides: Retention prediction. J. Chromatogr. A 2002, 958 (1–2), 167–182. DOI: 10.1016/S0021-9673(02)0 0306-0
(5) Lardeux, H.; Goyon, A.; Zhang, K.; et al. The impact of low adsorption surfaces for the analysis of DNA and RNA oligonucleotides. J. Chromatogr. A 2022, 1677, 463324. DOI: 10.1016/j.chroma.2022.463324
(6) Arend, K. Separation of siRNA Duplex and its Single Strands by YMC-Pack Diol SEC Columns, YMC Application Note, 2023.
(7) Deleavey, G.F.; Watts, J.K.; Alain, T.; et al. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acid Res. 2010, 38 (13), 4547– 4557. DOI: 10.1093/nar/gkq181
(8) Noll, B.; Seiffert, S.; Vornlocher, H.-P.; Roehl, I. Characterization of small interfering RNA by non-denaturing ion-pair reversed-phase liquid chromatography. J. Chromatogr. A 2011, 1218 (33), 5609–17. DOI: 10.1016/j.chroma.2011.06.057
(9) Eßer, D.; Rojahn, A.M. Improving Chromatographic Results for Oligonucleotides with Column Hardware. The Column 2023, 19 (2), 2–8.
(10) Beverly, M.; Hartsough, K.; Machemer, L. Liquid chromatography/electrospray mass spectrometric analysis of metabolites from an inhibitory RNA duplex. Rapid Commun. Mass Spectrom. 2005, 19, 1675–1682. DOI: 10.1002/rcm.1972
Ann Marie Rojahn studied chemistry and biotechnology in combination with an apprenticeship as a chemical laboratory assistant at the University of Applied Sciences in Krefeld, Germany. After receiving a bachelor’s degree, she pursued a master’s programme in chemistry at the University of Düsseldorf, Germany, with a focus on organic chemistry. Since 2019, she has worked for YMC Europe in Dinslaken, Germany, as a product specialist in the analytical chromatography team.
Kirstin Arend studied biology at the Ruhr University Bochum, Germany. After her master’s degree, she continued her studies at the Ruhr University Bochum and received her Ph.D. in microbiology in 2022, focusing on antibiotic research. In December 2022, she started to work at YMC Europe in Dinslaken as a trainee in the analytical chromatography team.
Daniel Eßer studied chemistry at the University of Applied Sciences Bonn Rhein-Sieg, in Rheinbach, Germany, with a focus on pharmaceutical and analytical chemistry. He received his Ph.D. in pharmaceutical and medicinal chemistry at the University of Düsseldorf, Germany. During his postdoc at the Institute of Pharmaceutical and Medicinal Chemistry of the University of Düsseldorf, he established a nanoLC–MS system. In 2013 he joined YMC Europe as a product specialist for analytical chromatography. Since 2017, he has been responsible for YMC’s analytical (U)HPLC column portfolio as product manager for analytical chromatography. Email: esser@ymc.de

New Study Reviews Chromatography Methods for Flavonoid Analysis
April 21st 2025Flavonoids are widely used metabolites that carry out various functions in different industries, such as food and cosmetics. Detecting, separating, and quantifying them in fruit species can be a complicated process.
University of Rouen-Normandy Scientists Explore Eco-Friendly Sampling Approach for GC-HRMS
April 17th 2025Root exudates—substances secreted by living plant roots—are challenging to sample, as they are typically extracted using artificial devices and can vary widely in both quantity and composition across plant species.