Validation and Peptide Mapping
LCGC North America
Peptide mapping is one of the preferred techniques for the comprehensive characterization of biopharmaceutical products and is often the analytical method of choice for studying a protein's primary structure.
Peptide mapping is a very common application in the biopharmaceutical laboratory and has become a true workhorse technique (1). It is used typically as an identity test for proteins, especially those obtained by recombinant DNA technology. A peptide map generated by a chemical or enzymatic treatment of a protein followed by a reproducible high-resolution chromatographic separation is capable of identifying single amino acid differences. This "fingerprint," sensitive to even the smallest change in a protein's structure, makes it an extremely valuable tool for identity testing and process monitoring. Because of this sensitivity, a peptide map can be used not just for the identification of proteins based upon the elution pattern of the peptide fragments in the separation, but also for the determination of posttranslational modifications, the confirmation of genetic stability, and the analysis of protein sequence when interfaced to a mass spectrometer.

Michael E. Swartz
While it is necessary to resolve each peptide fragment into a single peak, peptide mapping also represents a significant chromatographic challenge due to the large number of peptides that are generated from the enzymatic digest of a protein, and the significant number of alternative peptide structures (such as posttranslational modifications from proteolysis, phosphorylation, N-terminal acetylation and glycosylation, and oxidations) that also can be obtained.
Like any other analytical procedure, when used in a biopharmaceutical laboratory, the method used to generate or evaluate the peptide map must be validated.

Ira S. Krull
Applicable guidance is available from both the United States Pharmacopeia (USP) and the International Conference on Harmonization (ICH) on method validation in general (2,3). However, more specific USP guidance for peptide mapping validation also is available, and certainly should be examined for more detailed information (4). This column will highlight the USP guidance; however, for more information and background, the reader is encouraged to review the appropriate USP chapters.
The Biocharacterization "Map Quest"
Peptide mapping involves comparative testing of specific maps for each unique protein (the test sample) against a reference standard or reference material treated in an identical fashion. It is the end-product of one of several potential chemical processes that ultimately provide information about the protein under study. The process of generating a peptide map consists of four steps: isolation and purification of the protein, selective cleavage into the resulting peptides, the chromatographic separation, and the final analysis and identification of the peptides.
Isolation and purification are necessary for dosage forms or bulk drugs that might have excipients or additional active ingredients that can interfere with the protein of interest. When an isolation or purification step is employed, quantitative recovery should be validated against a reference standard.
Selective Cleavage of the Protein Peptide Bonds
The cleavage approach used is very dependent upon the protein test sample. Cleavage can be either enzymatic or chemical, and each type has multiple cleavage agents, as summarized in Table I. Complete cleavage is more likely to occur with enzymes compared to chemical agents. However, the overall goal is simply to have enough peptide fragments to be meaningful. If there are too many fragments, the map will lose its specificity and it could become more difficult to discern differences. The purity and activity of the cleavage agent also should be examined.
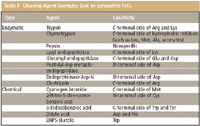
Table I: Cleaving Agent Examples (not an exhaustive list).
Sometimes pretreatment of either the sample or the cleavage agent is necessary. It also might be necessary to selectively protect the sample protein to guard against generating too many peptides, or it might be necessary to concentrate the protein or to separate it from added substances or stabilizers that can be used in some drug formulations. Sometimes chaotropic agents (for example, urea, guanidine, or surfactants) can be added to unfold the protein to allow full access to cleavage sites.
The cleavage process also has to be optimized, and the factors that affect the completeness and effectiveness of protein digestion are the same that would affect any chemical or enzymatic reaction. Temperature, pH, time, and the amount (enzyme/protein ratio) of cleavage reagent used are all important factors. Generally, a temperature between 25 °C and 37 °C is adequate for most digestions, and a pH is chosen appropriate for the cleavage agent, enzyme (will not denature the enzyme before it can react with the protein), and preservation of the integrity of the sample protein. Time can vary considerably, and if there is enough sample available, it is advisable to perform a time course study. The amount of cleavage agent used should be enough to accomplish a reasonably fast digestion time, but not so much that it interferes with the chromatographic map pattern. There is a compromise between using too much or too little of the cleavage agent, in order to avoid autodigesting the enzyme while generating an adequate map, and overdigesting and losing the "true" map and its information content.
Typically, a protein-to-protease ratio between 20:1 and 200:1 is used, and blank determinations also are made using a digestion control with all reagents included except for the protein of interest. Immobilized enzyme reagents also have been used to digest proteins for for peptide mapping, as a way to avoid autodigestion of the enzymes (5).
Chromatographic Separation
Many different techniques, as well as different modes of chromatography, are used to separate peptides for mapping. These include various forms of polyacrylamide gel electrophoresis (PAGE), capillary electrophoresis (CE), and reversed-phase liquid chromatography (LC), ion-exchange chromatography (IEC), and hydrophobic interaction chromatography (HIC).
Reversed-phase LC is arguably the most common technique employed, and the column, mobile phase, and gradient or (rarely) isocratic conditions used can be critical to the success of the separation.
Columns used for peptide maps are generally porous silica, 1.7–5.0 μm in size, with pore sizes ranging from 100 Å to 300 Å and ligands consisting of C18 (USP L1) or C8 (USP L7). Temperature control of the column is important for good repeatability. The most common mobile phases used consist of water and acetonitrile, with various additives, for example, trifluoroacetic acid or formic acid. If a buffer must be used, phosphate buffers provide the most flexibility for the selection of pH, although some thought must be given to alternative buffers if MS detection is used. Due to the complexity of the resulting sample, shallow gradient separations generally are recommended, with segments sometimes optimized using step functions or different slopes to give better resolution of important regions. Detection at low UV wavelengths, 200–230 nm, are typical due to limited chromophores. A chromatogram of a peptide map example is illustrated in Figure 1.
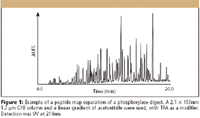
Figure 1
Validation of Peptide Mapping Methods
There are several critical factors that must be considered to validate a method used for peptide mapping, and each of the factors, along with the acceptance criteria, should be designed into a protocol or standard operating procedure (SOP).
The critical factors include robustness, the limit of detection, specificity, linearity, range, accuracy, and precision. Recovery and reagent stability are also important to consider during method validation. Recovery can be addressed by performing either quantitative amino acid analysis, spiking studies, or radiolabeling. Many of the validation parameters must not only address the separation, but also the fragmentation or digestion, particularly when considering robustness studies. The protocol also should include written test procedures that give a detailed description of the analytical method. Because there is a wealth of general validation guidance available (2–4), discussion will be restricted here to areas where peptide mapping validation might differ from other types of methods (for example, methods for synthetic drugs).
Robustness
To evaluate a peptide map against a standard, the chromatographic separation must be robust. Although general chromatographic robustness was covered in a previous "Validation Viewpoint" installment (6), the significance of pH and mobile phase composition should not be overlooked. However, there are additional issues to consider in a peptide map method, and these include (enzyme) reagent quality or purity, and digest stability
Column considerations also must be made during a proper robustness study, since it is a well known fact that no two chromatographic columns are created equal. When determining the robustness of the reagents used for digestion, it is common to evaluate a protein reference standard of known composition with cleavage agents from different lots. The number of peaks obtained, their shape, and the peak areas all are compared in the resulting chromatograms. Because in some cases, chromatographic run times can by quite long, the length of time and the conditions under which a digest can be stored before being analyzed also must be evaluated as part of a robustness study. Digest stability usually is evaluated by looking for significant differences in the map resulting from the analysis of several aliquots of a single digest stored at different conditions. It also can be desirable to investigate stability through to several freeze–thaw cycles.
Finally, it is a well known fact that no two chromatographic columns are created equal. Although manufacturers today have much better control of their processes than in the past, minor column differences can have a significant effect on the separation of these complex samples. It is a good idea to evaluate the reference standard on several different column lots, and to evaluate column life, because as a column ages, the separation can be affected. It is also a common precaution to examine alternative columns and to make slight modifications to the gradient profile where necessary to achieve equivalency.
Limit of Detection
The limit of detection (LOD) in a peptide map is determined by the ability of the method to distinguish changes in the map, for example, the presence or absence of a peak. Experiments can be carried out to modify the target protein intentionally, and then a digest of the modified protein is mixed with a control digest or standard reference material in varying proportions (7). Ideally, a decrease in peak response for the unmodified peptide and a corresponding increase for the modified peptide is observed. Peptides modified by oxidation, deamidation, or other mutations usually have reported LODs in the range of 2–15 mol% (7). The particular chemical modifications chosen to evaluate LOD should take into account both the protein and the expression host.
Precision
Precision in peptide mapping is measured on three levels; repeatability, and reproducibility from both intratest and intertest experiments. Repeatability is measured by running six replicate injections of a single pooled digest of the reference standard. When repeatability is performed in this manner, all variability from the sample and reagents are eliminated, and the true instrument or system component of precision can be measured and used to help set system suitability criteria.
Intra- and intertest measurements are the more important parameters to be evaluated during validation, however. Intratest precision is the reproducibility of the fragmentation (digestion) and the chromatographic separation. Acceptable precision is obtained when the peak retention times and areas are constant from chromatograms obtained from consecutive tests of a series of separately prepared digests of the test protein. The average standard deviation of the retention times and areas should not exceed a predetermined specified acceptance criterion.
Intertest precision is what traditionally has been referred to as intermediate precision or true reproducibility. It is a measure of the reproducibility of the peptide map when the analysis is run according to an experimental design made to measure the effects of the test run on different days, by different analysts, in different laboratories on different systems, different column lots. For intertest precision, the experimental design should include comparisons using peak retention times and areas relative to an internal standard peak within the same chromatogram. By using relative values, the need to make adjustments for things like injection volume differences, column volumes, and instrument gradient delay volumes is eliminated.
In general, it can be expected that %RSD for peak retention times and areas will be greater for the intertest compared with the intratest precision, which in turn will be greater than the repeatability results.
System Suitability
System suitability guidance can be found in the USP chapter on chromatography (8). Like any other method, the acceptance criteria for system suitability of a peptide map depend upon the identification of the critical test parameters that affect data interpretation and acceptance. System suitability limits (for both recovery and chromatography) are determined by running a reference standard in parallel with the test protein and looking for indicators that monitor, for example, that the desired endpoint was reached in the digestion (normally, selectivity and precision). However, the consistency of the pattern obtained is best defined by peak-to-peak resolution. Additional chromatographic parameters such as peak width, tailing factors, and column efficiency also can be used.
The parallel study (reference standard and test protein) also is used to visually compare each peaks relative retention time, responses (peak retention time and area), the number of peaks, and the overall elution pattern. This comparison often is complemented by mixing the two samples (1:1, v/v) and evaluating the peak response ratios and elution pattern. If all peaks in this mixed sample have the same relative retention times and peak response ratios, then the identity of the protein test sample can be confirmed. Significantly different retention times are also an indication of system variability, while the appearance of new or broader peaks indicates nonequivalence. Computer-aided pattern recognition software and other automated approaches have been used on occasion to examine the degree of difference or similarity when comparing two different peptide maps but have not gained routine acceptance.
Some Final Comments
At the Investigational New Drug (IND) phase, limited validation is necessary; typically only an approved test procedure that includes system suitability as a test control. Sometimes termed qualification, complete characterization of the individual peaks is not needed. As the regulatory process proceeds, a partial validation might be needed to give assurance that the method performs as intended in the development of a map for the test protein. However, validation of peptide mapping in support of further regulatory submissions requires a rigorous characterization of each of the individual peaks in the map. Methods that are used to characterize the peaks in a map commonly use mass spectrometry (MS).
In Figure 2, the LC–MS separation of a tryptic digest of α-1 acid glycoprotein, is an example of how MS can be used to characterize a peptide map. The MS detection was performed with a quadrupole time-of-flight (TOF) mass spectrometer, which is well suited for glycopeptides due to its extended mass range. Glycosylation is a posttranslational modification that plays a critical role in determining the efficacy and safety of a therapeutic protein. Glycosylation can be analyzed on the intact protein by MS, as released glycans by a variety of techniques, or as glycopeptides in LC–MS peptide maps. When glycosylation can be characterized with LC–MS of the glycopeptides, the site of attachment can be determined directly, and structural information can be obtained through MS-MS experiments. The example in Figure 2 shows that LC combined with electrospray ionization TOF-MS is a powerful tool for the rigorous characterization of peaks in a peptide map.
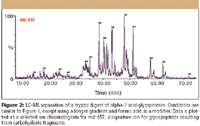
Figure 2
Conclusion
Although most of the underlying principles still apply, the validation of a peptide map includes some additional considerations when it comes to LOD, robustness, and precision, and depends upon the stage of the regulatory process. While mostly involving comparative testing, when properly validated, a peptide map can be used to accomplish its intended purposes: to confirm the primary structure of a protein, to detect whether or not alterations have occurred, and to demonstrate process consistency.
Michael E. Swartz
"Validation Viewpoint" Co-Editor Michael E. Swartz is Research Director at Synomics Pharmaceutical Services, Wareham, Massachusetts, and a member of LCGC's editorialadvisory board.
Ira S. Krull
"Validation Viewpoint" Co-Editor Ira S. Krull is an Associate Professor of chemistry at Northeastern University, Boston, Massachusetts, and a member of LCGC's editorial advisory board.
The columnists regret that time constraints prevent them from responding to individual reader queries. However, readers are welcome to submit specific questions and problems, which the columnists may address in future columns. Direct correspondence about this column to "Validation Viewpoint," LCGC, Woodbridge Corporate Plaza, 485 Route 1 South, Building F, First Floor, Iselin, NJ 08830, e-mail lcgcedit@lcgcmag.com
References
(1) B. Hancock, Ed., New Methods in Peptide Mapping for the Characterization of Proteins (CRC Press, Boca Raton, Florida, 1996).
(2) USP 29, 2006, Chapter 1225, p. 3050–3053.
(3) International Conference on Harmonization, Harmonized Tripartite Guideline, Validation of Analytical Procedures, Text and Methodology, Q2(R1), November 2005, see www.ICH.org.
(4) USP 29, 2006, Chapter 1047, p. 2875.
(5) G. Mark-Varga and P. Oroszlan, Eds., J. Chromatography Library 68, 188 (2003).
(6) M.E. Swartz and I.S. Krull, LCGC 24(5), 480 (2006).
(7) J. Bongers, J.J. Cummings, M.B. Ebert, M.M. Federici, L. Gledhill, D. Gulati, G.M. Hilliard, B.H. Jones, K.R. Lee, J. Mozdzanowski, M. Naimoli, and S. Burman, J. PBA 21, 1099 (2000).
(8) USP 29, 2006, Chapter 621, p. 2647.

University of Rouen-Normandy Scientists Explore Eco-Friendly Sampling Approach for GC-HRMS
April 17th 2025Root exudates—substances secreted by living plant roots—are challenging to sample, as they are typically extracted using artificial devices and can vary widely in both quantity and composition across plant species.













