The Use of Light-Scattering Detection with SEC and HPLC for Protein and Antibody Studies, Part II: Examples and Comparison to Mass Spectrometry
LCGC North America
Detailed examples of light-scattering techniques are presented as well as how their performance compares to MS and other methods.
Light scattering detection, particularly multiple-angle light scattering (MALS), can by very useful for the characterization of proteins and antibodies. In a number of applications, its performance exceeds that of mass spectrometry (MS) detection. In part I of this two-part article (1), we outlined the development of light-scattering detection in biotechnology, summarized its current uses and advantages, and explained the theory of how it works. Here in part II, we provide some detailed examples of its use and discuss how its performance compares to MS and other methods. As we will see in the examples, light-scattering analysis particularly excels at studying the bulk properties of proteins and antibodies in highly complex solutions.
Where Mass Spectrometry Falls Short, Light Scattering Steps In
When addressing the characterization of biopharmaceuticals or other complex biological samples, it is impossible to ignore analytical methods involving the combination of liquid chromatography and mass spectrometry (LC–MS) (2). These techniques range from high performance liquid chromatography (HPLC)–MS, where a relatively simple mixture is separated into several components and characterized by the intact masses and relative abundances (or even absolute concentrations, with the use of reference standards), to multidimensional LC techniques paired with high-resolution MS systems, which separate complex biological mixtures into thousands of peaks and perform in-depth analysis of individual species with multiple rounds of fragmentation. The information that is gained by in-depth LC–MS-MS analysis includes identification of post-translational modifications (PTM), oligosaccharide structure, and sequencing of peptides or even proteins with masses as large as 30 kDa.
In essence, LC–MS techniques are strongest for determining specific details about individual species. This ability is important for characterizing a substance with a limited number of variants, such as a small protein drug product. However, LC–MS falls short in quantitating the bulk properties of extremely complex substances, including PEGylated protein drug products (where PEG is polyethylene glycol), biologically derived substances such as low-molecular-weight heparin (LMWH), and inherently complex pharmaceutical substances such as copaxone. Light-scattering techniques pick up some slack in these areas.
Two additional challenges that cannot currently be addressed by LC–MS techniques are the determination of the tertiary or quaternary structure of a protein, and the study of protein–protein interactions (including aggregation). This information is irreversibly lost when proteins are ionized and vaporized by electrospray ionization or matrix-assisted laser desorption ionization (MALDI) for MS detection. Light-scattering techniques have the advantage of analyzing the protein in solution where the native conformations and protein–protein interactions are intact. In particular, size-exclusion chromatography (SEC)–MALS is especially valuable for very complex mixtures of proteins, antibodies, and aggregates (noncovalent or covalent), as well as highly derivatized proteins or antibodies as drug conjugates or PEGylated species.
Some Practical Biotechnology Applications of Light-Scattering Techniques
Below, we present several specific applications where light-scattering techniques adequately solve biotechnology challenges that are not currently fathomable by any LC–MS technique. All three involve studying highly complex solutions. Because many current protein-based pharmaceutical products are relatively large and complex, the development of therapeutic formulations at the high concentrations required can lead to a variety of complex protein stability issues (3). Many of these issues involve the interaction of protein therapeutics in vitro, leading to various degrees of mass-action associated and non-mass-action associated aggregation states.
Formulation Studies: Ensuring Protein Stability
A significant biotechnology application of light scattering technology is in formulation studies, investigating the stability of proteins in the solution used to deliver them to the patient. This can be accomplished by monitoring the protein's hydrodynamic radius (Rh), which will decrease or increase if the protein's native conformation collapses or expands because of disruption of stabilizing tertiary interactions. Figure 1 presents a study conducted using a Wyatt DynaPro plate reader, which simultaneously monitored an antibody formulation at a range of temperatures and various concentrations on a 384-well plate matrix (4). The Rh of the antibody is indicated as a function of temperature and concentration. The hydrodynamic radius exhibits a sigmoid relationship as a function of temperature for all antibody concentrations at pH 9.5. The change in midpoint temperature and radius as a function of concentration may indicate nonspecific attraction or oligomerization. Similar studies can be conducted with a range of pH levels and excipient concentrations to establish the most thermally stable formulation for any protein or antibody biopharmaceutical.
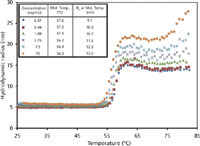
Figure 1: The hydrodynamic radius (Rh) of an antibody as a function of temperature and concentration.
Peptide mapping by reversed-phase ultrahigh-pressure liquid chromatography–mass spectrometry (UHPLC–MS) is an alternative approach to investigating protein stability. This technique specifically detects chemical degradations that involve a change in charge or mass, such as deamidation or oxidation of certain residues. However, this technique does not detect conformational changes that involve the physical rearrangement of the protein's three-dimensional structure. Therefore, using LC–MS alone, it is possible to miss protein population shifts from functional to nonfunctional states or noncovalent aggregation of denatured proteins. Conversely, a residue modification in the protein's active site may affect activity without resulting in a major change in conformation; in this case LC–MS may identify this substitution, whereas light-scattering techniques will not.
Formulation Studies: Minimizing Aggregration
Another key aspect of biopharmaceutical formulation is the prevention and detection of aggregation. Aggregation occurs if protein–protein interaction is thermodynamically favorable; this interaction varies with protein concentration and buffer pH, salinity, temperature, and overall composition. One measure of bulk protein–protein interaction in solution is the second virial coefficient (A2). The ability to rapidly and accurately measure this parameter aids in establishing the optimal buffer composition for drug products to mitigate aggregation. The automated SEC–MALS–refractive index (RI) method presented in Figures 2 and 3 is one such method of measuring A2. Light-scattering and refractive index measurements are obtained for each peak in a series of dilutions of one stock protein sample, and from this information the A2 coefficient is calculated for each peak (5). In Figure 2, the chromatogram in the upper right part of the figure shows the light-scattering signal of the 20–200 μL antibody solution injection series. The expanded boxed region shows the light-scattering signal in black and the differential RI signal in red, demonstrating complete dialysis of injected antibody with the mobile phase on the SEC column. Only the protein peaks are used in the on-line A2 analysis. Figure 3 shows a Zimm plot from the light scattering and concentration data of the protein peaks from Figure 2. Each color represents a different peak. From this Zimm plot, it can be determined that the antibody in phosphate-buffered saline (PBS) has an A2 of –5.46 × 10-5 mol mL/g2.
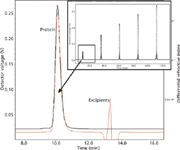
Figure 2: Light-scattering and refractive index measurement of serial injections.
Figure 4 shows another type of Zimm plot, perhaps the more customary format. Figure 4 illustrates an on-line A2 experiment to determine the molar mass, root mean square radius, and second virial coefficient of a sample (6). To do so, it is necessary to measure the light-scattering signal as a function of angle and concentration. The angular and concentration-dependent light-scattering data are then fit to the basic light-scattering equation using a Zimm plot (Figure 4, right). It is important to note that the global Zimm fit takes the data as a whole and extrapolation to zero angle or concentration is unnecessary. Together, these measurements yield insight into a number of properties of the protein and represent one of the most comprehensive determinations of the aggregation state.
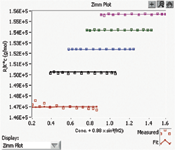
Figure 3: Zimm plot of an antibody in phosphate-buffered saline.
Using these methods, different buffer compositions can be sequentially analyzed to find the buffer that produces the optimal (small, positive) A2 coefficient. Another application for this method is in molecular biology, for finding the optimal conditions for protein crystallization that have a low, negative A2 coefficient.

Figure 4: On-line A2 analysis using a buffer-exchange column, with UV and DRI.
Figure 5 illustrates an SEC chromatogram in which the molar masses (y-axis) are plotted as the MALS detector 90° light-scattering signal versus the elution volume (x-axis), indicating the presence of a large range of molar masses (7). The peaks in this chromatogram are likely a result of different degrees of protein modifications, as well as aggregate formation, residual unreacted protein, and unreacted PEG, as indicated.
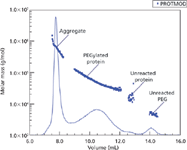
Figure 5: Plot of mass vs. volume from an SECâMALS analysis of a proteinâPEG complex.
Characterization of Low-Molecular-Weight Heparin by SEC–MALS
LMWH is manufactured by depolymerizing animal heparin, a heterogeneous substance, resulting in a mixture of linear polysaccharides of varying lengths. Heparin is made up of repeating disaccharide subunits that are sulfonated to various degrees. It is not a trivial challenge to characterize any individual species in the enormous multitude of structures that exist in this mixture. Sophisticated separation techniques would have to be used to isolate a single structure, and then multiple rounds of MS–MS may be necessary to unambiguously identify the linkages and levels of sulfonation of each saccharide of that single variant (although locations and anomericity of these modifications may be impossible to identify without nuclear magnetic resonance [NMR] spectroscopy). Biologically derived substances like LMWH are best characterized using bulk properties, and these features should be routinely examined between manufactured batches to ensure consistency. One of the most basic bulk features of a heterogeneous substance is the distribution of molar masses. Figure 6 shows how SEC–MALS was used to examine the molar mass distributions of three LMWH products generated by different depolymerization reactions. Three commercial LMWH products were examined by Scientific Protein Laboratories using a Waters 510 pump and 401 refractometer, a Shodex Ohpak SB-803HQ SEC analytical column (300 mm × 8.0 mm) with an SB-G guard column, and a Wyatt miniDawn triple-angle light-scattering instrument. The experiment was carried out in 0.1 M ammonium acetate buffer and dn/dc was measured off-line in a Wyatt Optilab DSP instrument operating at the same wavelength as the triple-angle instrument. The three LMWH products are distinguishable by their molar mass distributions (8).
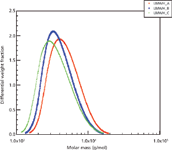
Figure 6: Molar mass distributions of three low-molecular-weight heparin products.
Molar mass distributions alone, however, are insufficient to characterize LMWH. Potency should be tested in vitro to confirm efficacy, and other key features that affect the activity of this substance, such as the concentration of the biologically active, unique pentasaccharide sequence that is known to interact with antithrombin, should be carefully monitored (9). Along with other analytical techniques, SEC–MALS is a key tool in characterizing LMWH and is used routinely in the quality control of these products.
Light Scattering Complements MS and Other Techniques
It is important to restate that the strength of light-scattering techniques is the ability to measure bulk properties of mixtures, such as the average molecular weight, the level of protein–protein interactions (A2), and hydrodynamic radius (Rh) and gyration radius or radius of gyration (Rg). When dealing with high-complexity mixtures like LMWH, these bulk properties provide the most useful information about the substance, rather than in-depth characterization of a few species.
On the other hand, light-scattering techniques do not provide great detail. For example, Rh is not as useful as complete protein structure by NMR, MS, or X-ray crystallography, and quantitation of PEGylation is not complete characterization, if the exact sites of PEGylation are not determined by LC–MS analysis. In summary, light-scattering techniques are essential for filling in the analytical gaps left behind by LC–MS techniques, and vice-versa.
Ensuring an Analytical Method Does Not Change the Nature of the Samples
When studying proteins in solution, it is also important that the analytical methods do not adversely affect the thermodynamics of the protein therapeutic so as to change the solution behavior during or after analysis. Because of its high degree of resolving power and relatively high-throughput nature, SEC has long been the industry standard for analyzing proteins in solution. This technique employs hydrostatic pressure along with a mechanical sieve to separate molecules based on their hydrodynamic characteristics. Although SEC has many advantages, there is concern over its analytical accuracy because proteins can interact with the stationary sieving matrix. It has been observed that the prolonged interaction between the proteins and the SEC matrix may lead to adsorption of the protein onto the column and possibly contribute to the artificial dissociation of preformed, weakly associated aggregates. In addition, the extent to which higher-order aggregates can permeate the pores of the stationary phase can also lead to an under-representation of these species. SEC also functions to dilute the injected sample, to varying degrees, which can also lead to disaggregation effects. Such defects in the SEC method itself are unwanted, because they change or alter the nature of the injected sample and distort the detector measurements, be these UV, refractive index, or light scattering.
It is therefore critical to use a first-principle technique to characterize and quantitate the protein samples eluted from the chromatography column, as well as techniques that do not involve mechanical sieving for preliminary separation. In other words, one should use direct MALS with no initial SEC or other separation step (as discussed further in the next section). MALS is one of the few first-principle techniques that can be used to determine the molecular weight and structure of proteins in solution, and it is applicable over the broadest range of any analytical method (10).
However, direct MALS or other direct light scattering methods, without SEC, are unable to determine how many dimers, trimers, or higher-order aggregates are present in the protein or antibody sample. Nor can they derive information about the size or shape (Rh or Rg) or A2 term of individual species in an unresolved mixture of such species.
Also, MALS and other light-scattering techniques produce a weight-averaged molecular weight and the corresponding z-average, square radii (r22)z (11,12), another reason why they also should be used after a separation method (such as SEC).
Using Light Scattering Without Chromatography in Free Solution
As mentioned above, to adequately characterize whether a method is changing the nature of the samples during characterization, it is essential to get a preliminary baseline of the free-solution nature of the protein complex (that is, without any initial separations step). For this measurement, another Rayleigh light scattering technique can be employed in a "batch" mode without using any separation mechanism. If the light-scattering intensity is measured over time, the resulting intensity fluctuations can be used to directly determine the translational diffusion coefficient (DT) for a protein, or protein complex in solution. Referred to as dynamic light scattering (DLS), this technique can be used to then convert the measured DT to a hydrodynamic radius (Rh) through the Stokes-Einstein equation. Although a more complete review of this technique is available through many sources, it is sufficient to say that this technique usually does not have the resolving power of SEC–MALS (2,12,13). Although a 3–5 fold difference in DT is required for resolution of species in solution when determining the mass-action aggregation of proteins, it is an invaluable tool when assessing the pre- and post-method, non-mass-action aggregation state. In Figure 7, the association state of an IgG1 antibody was measured using DLS at the time of preparation (pre-method) and after SEC analysis (post-method) (14). The DLS analysis clearly shows the degree of non-mass-action aggregation before analysis and the extent to which the SEC technique can artificially alter this aggregation state. The width of the histogram peaks represent the degree of polydispersity for each. Again, it must be understood that chromatographic conditions, even in SEC or gel permeation chromatography (GPC), can alter the nature of the original species injected onto the column, often in unknown ways. In this particular instance, the number of aggregates have decreased during the very method of analysis. Methods of analysis that change the original nature of a sample are meaningless to determine what was originally present in any such sample.
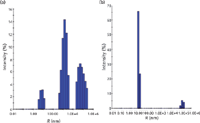
Figure 7: DLS analysis of pre- and post-method non-mass-action aggregation.
Limitations to SEC–MALS
Naturally, there are limitations to any analytical technique, and it is important to point these out when considering SEC–MALS characterization. Based on the theory of Rayleigh scattering, static light-scattering measurements can resolve the size of a protein complex (r2g)z down to about 1/20th of the incident wavelength (a radius of ~10 nm at 660 nm) and up to 1000 nm. Above 1000 nm, Rayleigh scattering does not hold, as the refractive index of the particles and that of the solvent produces a distortion of the electric field of the incident light that is solved only through the use of the Mie equation (2,12). As for molecular weight, for a given concentration c, the scattered light at zero angle is proportional to c multiplied by molecular weight. Thus, for protein samples with relatively low molecular weights (~1000 Da), relatively high concentrations are required to achieve a suitable signal-to-noise ratio. Alternatively, high-molecular-weight proteins and non-mass-action aggregates require very low concentrations to yield a suitable signal-to-noise ratio. This phenomenon raises the obvious issue of obtaining an adequate parallel mass concentration for each eluted fraction. For this reason, it is advisable to employ a differential refractometer detector as a universal concentration detector, along with a UV detector for its increased sensitivity, and selectivity, for low-protein-concentration species. These parallel concentration detectors can also provide valuable insight about the mass recovery of the injected protein samples onto the column and assist in the selection and conditioning of the stationary column matrix to minimize adsorption. A low mass recovery is not an acceptable set of SEC or GPC conditions for that particular analyte, and it should be rectified to ensure observing all the various species possible from a biotechnology product, adducts, aggregates, and derivatives. Provided that these conditions are achieved, SEC–MALS represents one of the most accurate and powerful tools available for characterizing the aggregation state of protein therapeutics in solution.
When to Use Mass Spectrometry and When to Use Light Scattering
In the preceding sections, we have described possible uses of SEC–MALS in the biotechnology industry, and where it can fill in some gaps left by SEC–MS methods. As already mentioned, MS is better for certain types of analysis and MALS is better for others. They are complementary techniques and should be used for those analyses for which they perform best or better than the other. Let us review some specific applications of each approach, especially MS, to highlight when to use each method (1).
Three Examples Where MS Works Best
Figure 8 first illustrates two SEC–UV–electrospray ionization (ESI)-MS chromatograms for a reduced and alkylated antibody, showing the heavy chain (HC), HC-HC dimer, and free light chain (LC) (15,16). The top chromatogram is the UV trace, and the bottom is the MS trace and spectra. Because the HC is a much higher molecular weight species (approximately 50 kDa), it shows a much more complex MS spectrum via ESI, with more multiply charged ions. Each of these individual peaks could be characterized by its molecular ion as well as by various fragmentation techniques involving high-resolution MS (such as Fourier transform MS), using collision-induced dissociation (CID) or electron-transfer dissociation (ETD) to sequence the amino acids and attachments (such as glycans), in each species. Because there are only three distinct species present and fully baseline resolved from one another, each of these can be readily, easily, and fully characterized. There is no need to use SEC–MALS on such species, even if the HC has several glycoforms present, as expected. Glycoprofiling can be performed easily by baseline-resolving the intact HC variants using other HPLC–MS conditions or by releasing all glycans and characterizing these, quantitatively and qualitatively, by conventional glycoprofiling using 2AB-tagging with hydrophilic interaction chromatography (HILIC), reversed-phase LC, capillary electrophoresis, and fluorescence detection (17,19). Problems with isobaric species in such studies do not generally arise, depending on the specific nature of the glycovariants. Between the high resolution obtained using UHPLC and the lack of isobaric variants in high-resolution MS, each such species should be identifiable. MALS will not provide any better characterizations for such species.
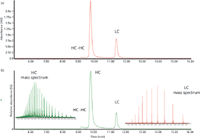
Figure 8: SECâUVâMS analysis of a reduced and alkylated monoclonal antibody, showing both UV and MS (TIC) chromatograms and mass spectra for both heavy chains (HC) and light chains (LC).
Figure 9 illustrates a slightly different scenario, somewhat artificially created, again for LC- and HC-derived species but now wherein the HCs are partially (not fully) alkylated on their residual thiol groups (15–17). This partial alkylation leads to a much larger number of HC species, in addition to glycovariants. Again, UHPLC conditions, whether with reversed-phase LC or ion-exchange chromatography (IEC), together with high-resolution MS, should be able to fully characterize each of these variants. It is, once again, not clear that MALS can play a significant role in adding to the characterization already possible using combined UHPLC–ESI-MS methods.
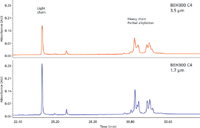
Figure 9: UHPLC separation of IgG heavy and light chains. Sample: Reduced and partially alkylated monoclonal antibody (IgG).
Figure 10 illustrates typical UHPLC–ESI-MS chromatograms for reduced monoclonal antibody (MAb) samples, comparing an innovator MAb to a generic or "biosimilar" product (19–20). The data were processed using Waters BiopharmaLynx software. In Figure 10a, we see that the light chain of the innovator and biosimilar MAb are identical. In Figure 10b, the heavy chain of the biosimilar candidate MAb shows a consistent mass shift of ~32 Da for all protein glycoforms compared to the reference sample (the innovator product). Glycosylation pattern differences were also detected. The molecular weight of each resolved species present can be accurately derived and sequenced, showing even very small differences between the HC species. Such results are clearly the forte of MS and not MALS, which is why it is important to recognize what each technique offers and is routinely capable of providing for a given analyte.
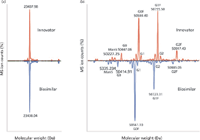
Figure 10: Typical UHPLCâMS chromatograms for reduced monoclonal antibody variants, comparing an innovator drug to a generic (biosimilar).
Where MALS Excels: Complex Mixtures
On the other hand, as we have described in the preceding sections of this column and in part I, MALS provides informational content for mixtures of protein- and antibody-derived species that are more complex than those described above, such as higher order aggregate species. (MS has problems, to unknown or undefined degrees, in determining any protein aggregate state, and surely not how much of each species is present.) MALS also provides Rh or Rg, unique numbers that indicate size, shape, and conformation for these species, and this is not possible by MS or other techniques now available. And, as the protein or antibody undergoes changes upon prolonged standing under stability testing conditions, under forced oxidation testing conditions, or on complexing with its antigen, MALS provides specific numbers that indicate changes in conformation and perhaps also changes in its biological activity. MS is not yet able to compete in such arenas. Such numbers become unique characteristics for identifying active versus inactive proteins or antibodies, and together with intact molecular weights obtained by MALS or MS, can be used to suggest retention or loss of bioactivity. Such information about size and shape is crucial information in the biopharmaceutical industry, not easily available by other, existing analytical techniques (such as surface plasmon resonance, enzyme-linked immunosorbent assay, or affinity capillary electrophoresis).
Summary
In summary, for easily resolved individual species, as shown in Figures 8–10, UHPLC–SEC–ESI-MS is perhaps the ideal approach for characterizing what species are really present. On the other hand, for more complex mixtures of proteins (synthetic or native), protein–antibody conjugates or aggregates (covalent or noncovalent), PEGylated proteins, antibody–drug conjugates, and related species, UHPLC–SEC–MALS appears to be the more advantageous analytical method.
Acknowledgments
This column has been a group effort, involving several unsung contributors, as well as those whose names appear on the first page. More specifically, we are indebted to several colleagues at Wyatt Technology in Santa Barbara, California, who read various sections, always making constructive and useful, suggested revisions. (Often, we actually listened to such suggestions.) In particular, our acknowledgement and appreciation goes to Phil Wyatt for the description of the fundamentals of light scattering theory and equations. Other people we wish to acknowledge at Wyatt include Cliff Wyatt, Geof Wyatt, Michelle Chen, and Sigrid Kuebler. Any errors of omission or commission are clearly those of the editors and coauthors alone.
References
(1) I.S. Krull, S. Kreimer, and A.S. Rathore, LCGC North Amer. 30(9), 842–849 (2012).
(2) D. Some and S. Kenrick, "Characterization of Protein–Protein Interactions via Static and Dynamic Light Scattering," in Protein Interactions, J. Cai and R.E. Wang, Eds. (InTech, Croatia, 2012, www.intechopen.com/download/pdf/32432), pp. 401–426.
(3) S.S. Shire, J. Pharm. Sci. 93, 1390–1402 (2004).
(4) Wyatt Technology Corporation, Application Notes, Thermal Stability as a Function of pH and Concentration, Santa Barbara, California, www.wyatt.com, 2012, Figure 2.
(5) Wyatt Technology Corporation, Application Notes, Automated, Online Second Virial Coefficient (A2) Measurements, Santa Barbara, California, www.wyatt.com, 2007, Figure 1.
(6) Wyatt Technology Corporation, Application Notes, Protein Solutions, Automated, Online Second Virial Coefficient (A2) Measurements, Santa Barbara, California, www.wyatt.com, 2007, Figure 2.
(7) Wyatt Technology Corporation, Application Notes, Protein Solutions, PEGylated Protein Modifications, Santa Barbara, California, www.wyatt.com, 1997, Figure 2.
(8) Wyatt Technology Corporation, Application Notes, Heparin Characterization, Santa Barbara, California, www.wyatt.com, 1997, Figure 2.
(9) J. Hirsh et al., Chest 119(1), 64S–94S (2001).
(10) P.J. Wyatt, Anal. Chim. Acta 272, 1–40 (1993).
(11) B.H. Zimm, J. Chem. Phys. 13(4), 141 (1945).
(12) C. Tanford, Physical Chemistry of Macromolecules (John Wiley & Sons, Inc., New York, New York, 1961).
(13) C.K. Matthews and K.E. van Holde, Biochemistry, Second Edition (The W.A. Benjamin/Cummings Publishing Company, Inc., Menlo Park, California, 1996).
(14) J. Champagne, unpublished study (2012).
(15) I.S. Krull, A. Rathore, and T.E. Wheat, LCGC North Amer. 29(12), 1052–1062 (2011).
(16) Waters Literature WA64266 and 720004076en (SEC), Waters Literature 720004018en (SEC-MS). Waters Corporation, Milford, Massachusetts.
(17) T. Wheat, Principles and Practice of UHPLC, UltraPerformance Now More Accessible Than Ever, training course, Waters Corporation (2010).
(18) A. Guttman, A. Rathore, and I.S. Krull, LCGC North Amer. 30(5), 412–421 (2012).
(19) M. Gilar, H. Xie, A.Chakraborty, J. Ahn, Y.Q. Yu, D.P. Dakshinamoorthy, W. Chen, St.J. Skilton, and J.R. Mazzeo, BioPharm Intl. 16–21 (August 2010 supplement).
(20) H. Xie, A. Chakraborty, J. Ahn, Y.Q. Yu, D.P.Dakshinamoorthy, M. Gilar, W. Chen, St.J. Skilton, and J.R. Mazzeo, mAbs 2(4), 379–394 (2010).
Simion Kreimer is a graduate student at Northeastern University pursuing a doctorate in the fields of chemistry and chemical biology through research in the Barnett Institute. His research interests include the application of MS and chromatography to translational medicine and pharmacology.

Simion Kreimer
John Champagne is a senior applications scientist and the Northeast regional manager for Wyatt Technology. He currently runs the applications laboratory for Wyatt Technology in the Boston area and provides both sample analysis and customer support services.

John Champagne
Ira S. Krull is Professor Emeritus of Chemistry and Chemical Biology at Northeastern University, Boston, Massachusetts, and a member of LCGC's editorial advisory board.

Ira S. Krull
Anurag S. Rathore is a biotech CMC consultant and an associate professor with the Department of Chemical Engineering at the Indian Institute of Delhi, India.

Anurag S. Rathore

Thermodynamic Insights into Organic Solvent Extraction for Chemical Analysis of Medical Devices
April 16th 2025A new study, published by a researcher from Chemical Characterization Solutions in Minnesota, explored a new approach for sample preparation for the chemical characterization of medical devices.
Sorbonne Researchers Develop Miniaturized GC Detector for VOC Analysis
April 16th 2025A team of scientists from the Paris university developed and optimized MAVERIC, a miniaturized and autonomous gas chromatography (GC) system coupled to a nano-gravimetric detector (NGD) based on a NEMS (nano-electromechanical-system) resonator.













