Ultrahigh-Performance Liquid Chromatography–Mass Spectrometry in Lipidomics
Special Issues
An analytical method for the global profiling of molecular lipids in biological samples, with particular emphasis on the plasmalogen lipids, is described.
This article describes an analytical method for the global profiling of molecular lipids in biological samples, with particular emphasis on the plasmalogen lipids. The global profiling method is based on ultrahighperformance liquid chromatography combined with quadrupole time-of-flight-mass spectrometry (UHPLC–QTOF-MS). The profiling approach is complemented by UHPLC–orbital trap MS in MSn mode for de novo lipid identification.
Lipids are an important class of essential metabolites and have many key biological functions. They are structural components of cell membranes, energy storage sources, and intermediates in signalling pathways (1,2). For example, tight control of membrane lipid composition is of central importance to maintain normal cellular physiology, and its dysregulation may affect membrane fluidity as well as topology, mobility, or activity of membrane-bound proteins. Lipids originate entirely or in part from two distinct types of building blocks: ketoacyl and isoprene groups. They are both functionally and structurally a very diverse group of compounds, partly because of the many possible variations of the lipid building blocks and the different ways of noncovalent linkage. The structural diversity of lipids is demonstrated by the huge number of molecular lipid species found In reply to: biological systems, which is estimated to be in the order of hundreds of thousands (3).
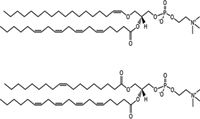
Figure 1: Structures of PC plasmalogen PC(p18:0/20:4) and corresponding PC(18:1/20:4).
Plasmalogens are a specific group of ether phospholipids. Structurally, they are glycerophospholipids which mainly consist of ethanolamine and choline, that is, phosphatidylcholines (PCs) and phosphatidylethanolamines (PEs) characterized by an alk-1'-enylether bond in position sn-1 where aliphatic moieties C16:0, C18:0, or C18:1 carbon chains are incorporated preferentially; the sn-2 position is specifically occupied by polyunsaturated fatty acids (Figure 1) (1–3). Plasmalogens are abundant lipid species representing 20% of the total pool of phospholipids in cells. They contribute to membrane structural integrity, and are also involved in multiple cellular functions, such as vesicle formation and membrane fusion (4–6), ion transport (7,8), and generation of secondary signal mediators, such as platelet activating factor (PAF) (9). The presence of the vinyl ether bond gives antioxidant properties to these molecules which diminishes free radical based cellular damage (10,11) and thus protects cells against oxidative stress. Indeed, plasmalogens have been shown to play an important role in human health as a factor involved in ageing, obesity, diabetes, and diseases of the central nervous system (12–14).
Global characterization of lipids in biological samples — lipidomics — is a challenging task because of the high diversity of lipids. These approaches include shotgun lipidomics, which uses direct infusion of lipid extracts into the mass spectrometer (15,16), as well as liquid chromatography coupled to mass spectrometry (LC–MS) (17). While the shotgun approach is relatively simple and rapid, it suffers from matrix effects and ion suppression, and it is not very well suited for global profiling of previously unknown lipids. The advantage of LC–MS-based methods over the shotgun approach is the higher sensitivity as well as the ability, by using a nontargeted strategy, to detect and identify novel lipids. However, matrix effects cannot be fully avoided in LC–MS either, and careful optimization of eluent composition as well as cleanup steps is required to avoid carry-over and contamination.
Here we describe an ultrahigh-performance liquid chromatography–mass spectrometry (UHPLC–MS)-based global lipidomics platform and identification workflow for the characterization of plasmalogens from biological samples. In the identification stage, an orbital trap MS system is used as the detector as this system provides outstanding mass accuracy and mass resolution. The ability to detect accurate masses allows the unequivocal compositional and structural elucidation of the compounds.
Experimental
Sample Preparation
A standard mixture 1 (20 µL) containing PC(17:0/0:0), PC(17:0/17:0), PE(17:0/17:0), and Cer(d18:1/17:0), (Avanti Polar Lipids, Inc.) and TG(17:0/17:0/17:0) (Larodan Fine Chemicals) was added to 10 µL of serum samples. HPLC-grade chloroform and methanol (2:1; 100 µL) were added to the samples, which were then vortexed for 2 min and allowed to stand for 30 min. Subsequently, samples were centrifuged and the lower phase (60 µL) was collected and 10 µL of internal standard mixture 2 was added. The internal standard mixture 2 contained the labeled lipids PC(16:1/0:0-D3), PC(16:1/16:1-D6), and TG(16:0/16:0/16:0-13C3).
The internal standard mixture contains compounds from several different lipid classes: PCs, PE, ceramide (Cer), phosphatidylserine (PS), phosphatidic acid (PA), as well as mono-, di- and triacylglycerols (MG, DG, and TG, respectively). Lipids are denoted by their molecular composition as follows: <Lipid class> <Number of carbon atoms in the first fatty acid moiety>:<Number of double bonds in the first fatty acid moiety> / < Number of carbon atoms in the second fatty acid moiety>:<Number of double bonds in the second fatty acid moiety>. For example, the abbreviation PC(17:0/17:0) indicates a phosphatidylcholine comprising two C17 fatty acids with no double bonds.
Instrumental Conditions
The extracts were analyzed on a Q-TOF Premier mass spectrometer (Waters, Milford) combined with an Acquity UPLC system (Waters) for the analysis of the whole data sets. For structural identification, UHPLC combined with TriVersa Nanomate (Advion Biosciences) electrospray ionization and LTQ-Orbitrap orbital trap MS (Thermo Fisher Scientific) systems was used, with the same analytical conditions for the UHPLC analysis. Separations were obtained using an 100 mm × 2.1 mm, 1.7-dp Acquity UPLC BEH C18 column (Waters) at a temperature of 50 °C. The solvent system included (A) ultrapure water (1% 1 M ammonium acetate, 0.1% formic acid) and (B) LC–MS-grade acetonitrile–isopropanol (1:1, 1% 1 M ammonium acetate, 0.1% formic acid). The gradient started from 65% A and 35% B, reached 80% B in 2 min, 100% B in 7 min, and remained there for 7 min. The flow rate was 0.400 mL/min and the injected volume was 2.0 µL (Acquity Sample Organizer, at 10 °C). Reserpine was used as the lock spray reference compound. The lipid profiling was performed using electrospray ionization (ESI) in positive mode and the data were collected at a mass range of m/z = 300–1200 with a scan duration of 0.2 s.
For the structural characterization with UHPLC–orbital trap MS, the following procedure was applied: Fractions collected from the UHPLC run were injected into the orbital trap mass spectrometer from the TriVersa Nanomate system using chipbased nanoelectrospray in positive and negative ion modes. Identifications were based on the exact mass (with target mass resolution of R = 60,000) and MSn spectra (LTQ). Plasmalogens and O-alkyl ethers were identified with the LTQ system in negative ion mode using normalized collision energy of 30%.
Data Processing
The data processing used MZmine 2 software (an open source software) (18) included alignment of peaks, peak integration, normalization, and peak identification. Lipids were identified using an internal spectral library. The data format specifies the identification number of the peaks, the average mass-to-charge ratio (m/z) value (average m/z), and the average retention time (RT) in seconds (average RT). The average values are obtained as the average value of the specific compound in the aligned dataset. The name of the compound is specified based on the search of the database.
Results
Lipidomic Analysis
The UHPLC–TOF-MS method covers the main classes of lipids, namely, cholesteryl esters (ChoE), PC, PE, Cer, MG, DG, TG, sphingomyelins (SM), and lysophosphatidylcholines (lysoPC). The sample preparation was based on modified Folch extraction, that is, liquid extraction using a mixture of chloroform:methanol (2:1), and these lipid classes are efficiently extracted by the method. For more acidic lipids, such as phosohatidylserines and phosphatidic acids, adjustment of the pH in the aqueous phase is required. The solvent system was chosen to be compatible with MS and to allow the ionization of a wide range of lipids. In addition, the solvent composition was optimized to contain a relatively strong solvent (acetonitrile–isopropanol) to obtain good peak shapes for late eluted TGs and also to avoid any significant carryover as a result of complex biological samples. Elevated temperature (50 °C) was also used to enhance the elution of the late eluted lipids. No significant carry-over was noticed, proved by blank analyses after real samples. The total analysis time for UHPLC–MS analysis was 12 min, thus allowing analysis of approximately 100 samples a day. Typically, around 800 lipid species can be detected from human plasma and serum samples.

Figure 2: Total ion chromatogram (ESI+) from UHPLCâTOF-MS lipidomics analysis of serum showing chromatographic separation of major lipid classes.
A typical LC–MS total ion current (TIC) chromatogram of a blood plasma sample obtained in positive ionization mode is shown in Figure 2. In the UHPLC separation, phospholipids are separated based on their fatty acid composition and degree of desaturation. Thus, lysophospholipids are eluted early, with lysoPCs containing one fatty acid attached to the glycerol backbone eluted before diacylphospholipids, which have two fatty acids. Phospholipids and sphingomyelins (SMs) are eluted in the same region while ceramides that do not contain a phosphatidylcholine (PC) head group as their sphingomyelin counterparts are eluted after PCs and SMs. In addition, DGs are eluted close to PCs and SMs. Cholesterol esters and TGs are eluted last, in the isocratic part of the gradient (100% of organic solvent), and the peaks are therefore slightly broader.

Table I: The calibration range and linearity of selected lipids
The quantitation of lipids was done by using groupspecific standards for the normalization of the data. All monoacyl lipids except cholesterol esters, such as monoacylglycerols and monoacylglycerophospholipids, were normalized with PC(17:0/0:0); all diacyl lipids except ethanolamine phospholipids were normalized with PC(17:0/17:0); all ceramides with Cer(d18:1/17:0); all diacyl ethanolamine phospholipids with PE(17:0/17:0); and TG and cholesterol esters were normalized with TG(17:0/17:0/17:0). Other (unidentified) molecular species were normalized with PC(17:0/0:0) for retention time <300 s, PC(17:0/17:0) for RT between 300 s and 410 s, and TG(17:0/17:0/17:0) for higher retention times.
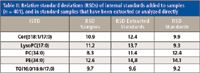
Table II: Relative standard deviations (RSDs) of internal standards added to samples (n = 401), and in standard samples that have been extracted or analyzed directly
The method was also linear over a large concentration range for most of the lipid classes (Table I). To control the quality of the lipid analyses, a set of control serum samples, standards extracted with the same protocol as samples and pure standards were analyzed together with the samples. The repeatability of the analysis was proved to be good, as shown in Tables II and III, with the relative standard deviations (RSDs) in internal standards added to samples (n = 401) below 12.2% which was the same level for the extracted standards. The instrumental variation was also in the same range.
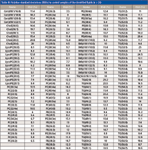
Table III: Relative standard deviations (RSDs) in control samples of the identified lipids (n = 26)
Plasmalogen Identification
In the UHPLC separation, plasmalogens were also easily separated from their acyl-lipid counterparts in the LC. A comprehensive set of MSn studies was performed for the identification of fatty acid composition in lipids and to differentiate between O-alkyl ethers and plasmalogens. In particular, ESI– was used for the identification of specific product ions for PE and PC plasmalogens. A spectral database, containing RT information as well, has been constructed based on these studies, using both standard compounds (limited availability), and the identification of specific plasmalogens and alkylethers in a variety of biological samples (serum/plasma and tissues).

Figure 3: ESI+ identification of two unknown lipids. (a) Fragmentation of [M+H]+ ion of a phosphatidylcholine produce an abundant ion at m/z = 184 corresponding to phosphocholine head group. (b) Neutral loss of phosphoethanolamine head group (141 u) is observed when [M+H]+ ion of a phosphatidylethanolamine is fragmented.
In the identification of lipids, and in particular plasmalogens, MSn was utilized, using both ESI+ and ESI–. In ESI+, only limited information could be obtained (Figure 3), mainly from the polar head group of the lipid (19). Therefore, ESI– mode is also needed (Figure 4) for detailed identification of the lipid structure and of the fatty acid side chains (20,21). In negative ion mode, PCs are detected mainly as formate adducts [M+COO]–, which will fragment to yield abundant [M–15]– ion. Fragmentation of [M–15]– ion (MS3) reveals information about the fatty acid composition of the PC lipids. The most abundant negative ions in MS3 spectra correspond to carboxylate anion from sn-2 and sn-1 position (R2 and R1 in Figure 4). In addition, MS3 spectra show a complementary ion which comprises fatty acyl (from sn-1 position) and glycerophosphocholine head group [M–15–R2COOH]–. MS4 analysis of [M–15–R2COOH]– ions reveals critical information regarding the two etherlipid subclasses, plasmalogens, and O-alkyl ethers (Figure 5). While the plasmalogen PC species, for example, produces a class specific ion at m/z 224, the O-alkyl-ethers instead yield a phosphatidic acid anion [M–15–R2COOH-89]– (as a result of the neutral loss of dimethylamino ethylene). Therefore, all three subclasses of important membrane lipids, plasmalogens, and O-alkyl ethers as well as diacyl PCs, can readily be distinguished using ESI– and MSn when the components are chromatographically separated. In a similar manner, the PE ether lipid subclasses can also be characterized based on unique fragments in MS3 spectra (22).

Figure 4: Fatty acid composition phospholipids can be characterized using ESIâ and fragmentation in ion trap. Fragmentation of PC(17:0/17:0) is shown as an example.
Conclusions
A UHPLC–MS-based lipidomics platform that uses high-resolution MS with both ESI+ and ESI– analysis and MSn fragmentation provides a powerful tool to characterize lipid subclasses. Phosholipids with ester linkages (diacyl components) can be identified based on accurate mass and chromatographic retention time, and the fatty acyl composition can be revealed based on MSn fragmentation of negative ions. In addition, MSn analysis in negative ion mode can readily distinguish the two etherlipid subclasses of phospholipids, namely plasmalogens and O-alkyl ethers. Plasmalogens produce a class specific ion at m/z 224 while the other ether phospholipids yield a phosphatidic acid.

Figure 5: An example of fragmentation of a plasmalogen- and O-alkyl ether PCs: (a) MS4 analysis of plasmalogen PC(p18:0/20:4) produce two complementary fragments. Negative ion at m/z = 224 is a specific fragment for plasmalogen PC species. (b) MS4 spectra of O-alkyl ether PC(O-16:0/16:0) shows instead only a phosphatidic acid anion.
Heli Nygren, PhD, is a senior scientist in the metabolomics laboratory at VTT Technical Research Centre of Finland. Her research activities focus on the development and use of advanced chromatographic and mass spectrometric methods for metabolomics.
Päivi Pöhö, MSc, is a scientist in the metabolomics laboratory at VTT Technical Research Centre of Finland. Her main field of research is lipidomics utilizing chromatographic and mass spectrometric methods.
Tuulikki Seppänen-Laakso, PhD, is a senior scientist at VTT Technical Research Centre of Finland. Her research activities include applications of lipidomics in life sciences.
Ulla Lahtinen is a senior technician in the metabolomics laboratory at VTT Technical Research Centre of Finland. Her work is focused on the sample preparation of biological samples as well as lipidomics with hyphenated chromatographic methods.
Matej Orešic holds a PhD in biophysics from Cornell University in Ithaca, New York. Since 2003 he has led the research in domains of quantitative biology and bioinformatics at VTT Technical Research Centre of Finland, where he is a research professor in systems biology and bioinformatics. Professor Orešic is a director of the Academy of Finland Centre of Excellence in Molecular Systems Immunology and Physiology Research. His main research areas are metabolomics applications in biomedical research and integrative bioinformatics.
Tuulia Hyötyläinen, PhD, leads the metabolomics team at the VTT Technical Research Centre of Finland. Her research is focused on chromatographic and mass spectrometric techniques applied to metabolomic studies. Her main research centres on the development of methodologies for comprehensive chemical characterization of complex biological samples, from sample pretreatment to data analysis and identification of novel metabolites.
References
(1) M. Orešic, V. Hänninen, and A. Vidal-Puig, Trends Biotechnol. 26(12), 647–652 (2008).
(2) E. Fahy et al., J Lipid Res. 46(5), 839–861 (2005).
(3) L. Yetukuri et al., Molec BioSyst. 4(2), 121–127 (2008).
(4) W.C. Breckenridge et al., Biochim Biophys Acta 320(3), 681–686 (1973).
(5) K. Lohner et al., Biochim Biophys Acta 1061(2), 132–140 (1991).
(6) P.E. Glaser and R.W. Gross, Biochemistry 34(38), 193–203 (1995).
(7) R.J. Bick et al., Arch Biochem Biophys. 286(2), 346–352 (1991).
(8) D.A. Ford and C.C Hale, FEBS Lett. 394(1), 99–102 (1996).
(9) W. Chao and M.S. Olson, Biochem J. 292(3), 617–629 (1993).
(10) B. Kuczynski and R.V. Reo, Neurochem Res. 31(5), 639–656 (2006).
(11) B. Engelmann et al., Biochem Biophys Res Commun., 204(3), 1235–1242 (1994).
(12) R. Maeba et al., J Atheroscler Thromb. 14(1), 12–18 (2007).
(13) X. Han et al., J Neurochem. 77(4), 1168–1180 (2001).
(14) K. H. Pietiläinen et al., PLoS One 2(2), e218 (2007).
(15) S. Moco et al., TrAC. 26(9), 855–866 (2007).
(16) H.R. Jung et al., Biochim Biophys Acta 1811(11), 925–934 (2011).
(17) H. Nygren et al., Methods Mol Biol., 708, 247–257 (2011).
(18) T. Pluskal et al., BMC Bioinformatics 11(1), 395 (2010).
(19) F.F. Hsu and J. Turk, J Am Soc Mass Spectrom. 14(4), 352–363 (2003).
(20) N. Khaselev and R.C. Murphy, J Am Soc Mass Spectrom. 11(4), 283–291 (2000).
(21) F.F. Hsu and J. Turk, J Am Soc Mass Spectrom. 18(11), 2065–2073 (2007).
(22) K.A. Zemski Berry and R.C. Murphy, J Am Soc Mass Spectrom 15(10), 1499–1508 (2004).

Determining Enhanced Sensitivity to Odors due to Anxiety-Associated Chemosignals with GC
May 8th 2025Based on their hypothesis that smelling anxiety chemosignals can, like visual anxiety induction, lead to an increase in odor sensitivity, a joint study between the University of Erlangen-Nuremberg (Erlangen, Germany) and the Fraunhofer Institute for Process Engineering and Packaging (Freising, Germany) combined behavioral experiments, odor profile analysis by a trained panel, and instrumental analysis of odorants (gas chromatography-olfactometry) and volatiles (gas chromatography-mass spectrometry).
Investigating 3D-Printable Stationary Phases in Liquid Chromatography
May 7th 20253D printing technology has potential in chromatography, but a major challenge is developing materials with both high porosity and robust mechanical properties. Recently, scientists compared the separation performances of eight different 3D printable stationary phases.
Detecting Hyper-Fast Chromatographic Peaks Using Ion Mobility Spectrometry
May 6th 2025Ion mobility spectrometers can detect trace compounds quickly, though they can face various issues with detecting certain peaks. University of Hannover scientists created a new system for resolving hyper-fast gas chromatography (GC) peaks.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)










