Simple and Rapid MRM Method Development for Analyzing 288 Pesticides by GC–MS-MS
Gas chromatography coupled with tandem mass spectrometry (GC–MS-MS) operated in MRM mode is a powerful technology for multi-residue analysis in complex food matrices. Good GC separation combined with MS-MS selectivity facilitates monitoring hundreds of MRM transitions within a short cycle time which allows for analysis of large number of compounds in complex food matrices. Nevertheless, MRM method development is time consuming and labor-intensive, which mainly comes from mapping retention time, MRM transition set-up and post-data processing.
Gas chromatography coupled with tandem mass spectrometry (GC–MS-MS) operated in MRM mode is a powerful technology for multi-residue analysis in complex food matrices. Good GC separation combined with MS-MS selectivity facilitates monitoring hundreds of MRM transitions within a short cycle time which allows for analysis of large number of compounds in complex food matrices. Nevertheless, MRM method development is time consuming and labor-intensive, which mainly comes from mapping retention time, MRM transition set-up and post-data processing.
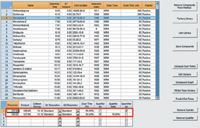
Figure 1: The CBS acquisition method table with MRM transitions linked to each pesticide.
In this study, an innovative software feature, compound-based screening (CBS), is introduced on a Bruker SCIONTM TQ GC–MS-MS system for method development of 288 pesticide analysis. CBS is implemented with a built-in factory compound library containing optimized MRM transitions along with retention indices for hundreds of pesticides. With retention index (RI), retention time (RT) can be predicted from a small subgroup of pesticides from an initial run within a narrow retention time window, saving time and effort for searching the retention time over the entire run for each pesticide. After obtaining retention time, the scan time (dwell time) for each MRM can be optimized automatically based on the average peak width and desired scan points across each peak. Furthermore, the acquisition and data processing section are integrated within the new platform so that changes on either section can be updated automatically.
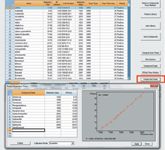
Figure 2: Prediction of retention time for all target pesticides from a subgroup of pesticides.
Experimental
Sample:
The standard pesticide mix was prepared by spiking 288 standard pesticides in 1:1 hexane/acetone solvent.
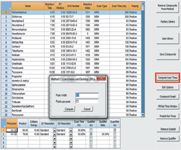
Figure 3: Optimization of dwell time for timed MRM.
Instrument:
SCIONTM TQ triple quadrupole mass spectrometer coupled to a Bruker 451 GC and CP 8400 Autosampler
GC-MS conditions:
Column: BR-5ms 30 m × 0.25 mm i.d. and 0.25 µm
Carrier gas: He at 1 mL/min
Oven: 80 °C (2 min), at 20 °C/min to 160 °C , at 5 °C /min to 280 °C, at 20 °C/min to 300 °C(5 min), total 36 min
Injection: Splitless, 280 °C, 1 µL
EI source Temp.: 260 °C
Transfer line Temp.: 300 °C
Emission Current: 80 µA
Q2: Ar (1.5 mTorr)

Figure 4: Timed MRM windows for 288 pesticide analysis.
Results and Discussions
A factory-supplied compound library with optimized MRM transitions and retention indices has been implemented in Bruker MSWS 8, the data system for SCION TQ. More than 800 compounds, including more than 400 pesticides, are included in the library. The following illustrates the five-step quick approach for setting up an MRM method for 288 pesticides:
1) Select the 288 target pesticides in the mixture by name or CAS # from factory library to export to method table, the MRM table will be automatically populated (Figure 1).
2) Run a full scan of the pesticide mixture standard. Pick up 12 pesticides with identified retention times across the entire run. Click the Predict RT function, select to input this subset of pesticides with RT and RI, plot the RT as a function of RI and to select the best fit, click Apply to input the predicated retention times for all pesticides in the method acquisition table (Figure 2).
3) In the next couple of runs, locate the RT for the rest of pesticides as well as refine those RT that found in above step.
4) Compute the scan (dwell) time for MRM transitions of each individual pesticide by clicking "Compute Scan Time" button, the scan time for each MRM transitions/compound is automatically assigned with the optimized scan (dwell) time to schedule a timed MRM approach (Figure 3).
5) Update the method with the refined RT information. The method is ready for running calibration curves (Figure 4).
Statistical analysis of predicted RT vs. experimental RT was carried out, and the result listed in Figure 5. As shown, 44% of 288 compounds are within ±0.5 min window from the predicted RT; 27% within a ±0.5–1 min window. A total of 91% pesticides fall within a ±2.5 min window, demonstrating adequate accuracy of the prediction model. The RT prediction function significantly simplifies MRM method development process for multi-residue analysis applications, which ultimately improves the work efficiency in the production lab.

Figure 5: Statistics of predicted retention time vs. real retention time of 288 compounds.
Conclusions
Compound based screening (CBS) along with the factory built-in MRM library, retention time prediction, and dwell time optimization features in Bruker MSWS 8 significantly simplifies MRM method development for multi-residue analysis.
Bruker Daltonics
Billerica, MA
tel. (978) 667-3660, fax (978) 667-5993
Website: www.bruker.com

Separation of Ultra-Short and Long Chain PFAS Compounds Using a Positive Charge Surface Column
December 11th 2024A separation of ultra-short and long chain PFAS (C1-C18) is performed on a HALO®PCS Phenyl-Hexyl column along with a HALO®PFAS Delay column which demonstrates excellent retention for both hydrophilic and hydrophobic analytes.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)














