Pesticide Analysis in Dietary Supplements Using Gas Chromatography Time-of-Flight Mass Spectrometry
The Application Notebook
Screening for pesticide content is a common analysis that is done in laboratories where many samples need to be analyzed over a short period of time. The desire for a high-speed analysis is due to the need for a fast result from a potentially large volume of samples. A fast analysis is typically achieved through the use of high carrier gas flow rates in combination with temperature programming. A fast detector is needed in order to fully characterize the narrow chromatographic peaks generated from these experiments. TOFMS is ideal for detecting these narrow peaks because of its fast acquisition rate over a full mass range at all times during the experiment. In addition, TOFMS provides for non-skewed mass spectra making peak deconvolution possible.
Megan McGuigan, LECO Corporation
Screening for pesticide content is a common analysis that is done in laboratories where many samples need to be analyzed over a short period of time. The desire for a high-speed analysis is due to the need for a fast result from a potentially large volume of samples. A fast analysis is typically achieved through the use of high carrier gas flow rates in combination with temperature programming. A fast detector is needed in order to fully characterize the narrow chromatographic peaks generated from these experiments. TOFMS is ideal for detecting these narrow peaks because of its fast acquisition rate over a full mass range at all times during the experiment. In addition, TOFMS provides for non-skewed mass spectra making peak deconvolution possible.

In this report, a screening method for Organochlorine (OC) and Organophosphorus (OP) pesticides is discussed. Initial method development experiments were done with a standard pesticide mixture (Restek). Extracts of Ginkgo Biloba and Milk Thistle were then studied to determine their possible pesticide content with a total experimental run time of ~22 min.
Results and Conclusions
Figure 1 shows an Analytical Ion Chromatogram (AIC) for the separation of 43 OC and OP pesticides. Shown in the inset is a detailed view of the chromatographic overlap of chlorpyrifos and parathion plotted as the AIC along with m/z 197 and 291. The AIC trace appears to show just a single peak, however the automatic peak find and deconvolution features in ChromaTOF identified two target pesticides under that peak. On either side of the inset of Figure 1 are the Caliper, Peak True (deconvoluted) and Library Hit spectra for each of the overlapping components. The Caliper spectrum contains all ions present at the peak marker. Following deconvolution, ions from overlapping peaks are removed resulting in the Peak True spectrum. Peak identifications are then made by comparing the Peak True spectrum with the NIST library. Following the identification of all the target analytes in the standard mixture, the standard was used as a reference (single-point calibration) in order to identify any target pesticides in the supplement. Constraints were set on the retention time, minimum s/n threshold and spectral similarity. The use of the reference allows for the automated identification of target analytes in subsequent samples.
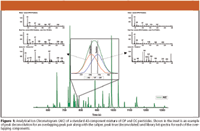
Figure 1
Figure 2 shows shows chromatograms for the analysis of extracts from Gingko Biloba (a) and Milk Thistle (b) plotted as the AIC. With a s/n threshold of 50, a total of 223 compounds were identified in the Gingko Biloba extract and 88 compounds were identified in the Milk Thistle extract. In both samples, the only pesticide that was identified was Dieldrin, which is highlighted in both chromatograms. The corresponding dieldrin mass spectra (deconvoluted peak true) are shown in the inset, along with the reference spectrum.
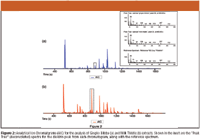
Figure 2
The use of fast GC conditions combined with high-speed data acquisition, automated data processing and reference chromatogram comparison resulted in a fast turn-around for sample analysis. By taking advantage of the automated peak find and deconvolution software features, complete chromatographic resolution is not necessary.

LECO Corporation
3000 Lakeview Avenue, St. Joseph, MI 49085
tel. (269) 985-5496, fax (269) 982-8977
















