Nontargeted Metabolite Profiling by UHPLC–MS: A Case Study in Beer Reveals Effect of Temperature on Nonvolatile Small Molecules During Storage
LCGC North America
This analytical approach can be applied in many fields; the example illustrates its use in a food chemistry application.
Nontargeted metabolite profiling by ultrahigh-pressure liquid chromatography (UHPLC)coupled with mass spectrometry (MS) is a powerful technique to investigate metabolism. The approach offers an unbiased and in-depth analysis that can enable the development of diagnostic tests and novel therapies, and further our understanding of disease processes. Other fields such as ecology, plant biology, and food chemistry can also benefit from this approach. Here, the power of nontargeted metabolite profiling is illustrated in a study focused on assessing the impact of storage conditions on flavor stability in beer.
The term metabolomics broadly describes an approach rather than a particular technique. For example, choosing among a variety of instrument platforms, you can perform a metabolomics experiment in a targeted or nontargeted manner or a combination of both (1). Historically, metabolite analysis was performed in a targeted manner focusing on a few known metabolites important to the biological question at hand. In such experiments, the biological hypothesis dictates the choice of metabolites targeted, and the analytical steps are optimized to detect those molecules. In recent years, metabolite research has transitioned to a more "omics" approach. Thus, experiments are performed in a broad and unbiased manner, enabling the detection of as many metabolites as possible. The results from a nontargeted metabolomics experiment can generate new hypotheses and drive the next step of research, which often involves switching to a targeted metabolomics workflow. It is also possible — as well as powerful — to combine the two approaches, performing the experiment in a nontargeted manner while at the same time monitoring a panel of known molecules.
State-of-the-art mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy are the most frequently used techniques for nontargeted metabolite profiling. NMR excels at identifying unknown compounds. Its applicability is limited, however, because it requires large sample volumes and high concentrations, is a lower throughput technique, and has a narrow depth of coverage (2). MS offers the significant advantages of high-throughput analysis, minimal sample requirements (microliter volumes), and a broad depth of coverage. Yet its ability to confidently identify and confirm unknown compounds is substantially limited.
Structural elucidation of small molecules by MS is accomplished through the acquisition of tandem mass spectra (MS-MS), which involves the fragmentation of ionized molecules within the mass spectrometer. Small-molecule spectral libraries are well developed for gas chromatography–mass spectrometry (GC–MS) applications utilizing electron ionization, but have been slow to develop for liquid chromatography–mass spectrometry (LC–MS) applications. Unlike peptides, which fragment predictably, small-molecule metabolites are structurally diverse, making their behavior in the mass spectrometer less predictable. This unpredictability has greatly inhibited the generation of theoretical spectral databases as used in proteomic applications (3). Recently, some progress has been made in generating small-molecule LC–MS spectral libraries through the analysis of authentic standards (4,5), and these libraries continue to grow. However, given the complexity of the metabolome, its variation among biological species, and the variation in molecular fragmentation with different MS conditions, generating comprehensive spectral libraries through the analysis of authentic standards is a slow and tedious process. For those reasons, the process of molecular identification remains a significant bottleneck in nontargeted, LC–MS-based metabolite profiling studies (6,7).
Most MS-based, nontargeted metabolite-profiling workflows exploit the fast separations and narrow peak widths obtained by coupling a mass spectrometer with an ultrahigh-pressure liquid chromatography (UHPLC) system. Traditionally, in such a workflow (see Figure 1), only the MS signal (that is, no fragmentation) is collected for each detected molecular feature, optimizing the number of points per peak over the narrow UHPLC elution profiles. This approach ensures the validity of the quantitative data obtained for each feature. The quantitative data are used in downstream statistical analysis to determine molecular features of interest (such as potential biomarkers). Molecular identification is then attempted using a combination of accurate-mass measurements, isotopic distribution, and additional experimental sample analysis to obtain the MS-MS fragmentation spectrum for each feature of interest. The accurate mass and isotopic distribution can be used to predict the compound's molecular formula. The experimental MS-MS spectrum can be used to infer structural information or, in the best-case scenario, to compare with the MS-MS spectrum of an authentic standard (1,7).

Figure 1: UHPLCâMS-based nontargeted metabolite profiling workflow. Compound identification involves the targeted acquisition of MS-MS spectra for compounds of interest (for example, potential biomarkers) and comparison with authentic standards.
Nontargeted metabolomic profiling offers an in-depth biochemical analysis applicable to any biological organism or tissue. For example, it is used in many studies that focus on human health to advance our understanding of disease processes and ultimately to enable the development of diagnostic tests and novel therapies. Indeed, the National Institutes of Health (NIH) recognizes the potential impact of the metabolomics field and has shown its commitment to metabolomics by recently establishing the Common Fund Metabolomics Program.
Nontargeted metabolomics can be applied in other fields that may be less obvious targets for this application. For example, a recent ecological study pursued a nontargeted metabolite-profiling approach to show that the rate of decomposition for leaf litter varied according to the decomposer microbial community. The finding could have important implications in the rate and efficiency of the formation of organic matter in soil and carbon sequestration in soil (8).
Food chemistry represents another field in which the nontargeted approach has been successfully applied (9). We recently presented the results of a study that adopted this approach to analyze beer. The goal of the study was to assess the effect on flavor stability of storage length and conditions, particularly as they relate to the nonvolatile small-molecule profile (10).
Why Study Beer with Non-targeted Metabolite Profiling?
An important metric in the brewing industry is "beer quality," defined by attributes of beer such as flavor, mouth-feel, aroma, and appearance. Unfortunately, storage conditions (including time, temperature, and light exposure) can influence beer quality and result in a limited shelf life for commercial products, after which they are no longer considered true to brand. A better understanding of the molecular cause and effect of the aging process can lead to improved brewing methods that will increase beer stability over a longer time and range of storage conditions.
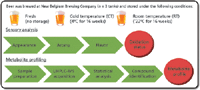
Figure 2: Experimental design.
Traditionally, analytical work in the brewing industry has focused on volatile compounds known to influence flavor and taste. Yet nonvolatile compounds can also be factors that contribute to — or inhibit — the staling process. In our study, we used a standard nontargeted UHPLC–MS metabolite-profiling workflow to determine whether changes in nonvolatile metabolites during storage could reflect beer quality. Thus, this study incorporated both a molecular-level analysis and a qualitative, sensory analysis (Figure 2).

Figure 3: Quantitative scale used in sensory analysis to score "oxidation" of beer.
Sensory Analysis of Beer
Sensory analysis was performed at New Belgium Brewery, where employees are expertly trained to qualitatively assess quality traits such as appearance, aroma, and flavor. A semiquantitative scale was used to determine an "oxidation score" according to perceived notes in the beer: sulfur, roasted malt, paper or cardboard, mold, bitter, caramel, bread, vinous, fruit, acid, cereal, resin, honey, waxy, smoke flavor, and altered mouth feel (Figure 3). The results of the sensory analysis indicated significant "oxidation" in beers stored both at room and cold temperatures, with an increased rate of oxidation at room temperature (Figure 4).
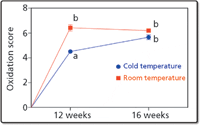
Figure 4: Results of sensory analysis illustrating significant perceived "oxidation" by an expert taste panel. Adapted from Heubeger and colleagues (10).
Nontargeted Metabolite Profiling of Beer
Nontargeted metabolite profiling was performed to determine the molecular signature representing the oxidation detected in the sensory analysis (Figure 5). For this experiment, sample preparation consisted only of a simple centrifuge step to remove particulates (although a protein removal step is typically applied in metabolomics studies involving the analysis of biological samples [11]). The beer metabolites were separated in time using a reversed-phase C8 column on a UHPLC system. As mentioned above, the use of UHPLC enables extremely fast and highly reproducible separations. Mass spectra were collected using a high mass accuracy, quadrupole time-of-flight (QTOF) instrument.
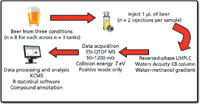
Figure 5: UHPLCâMS nontargeted metabolite-profiling workflow
This experiment involved the analysis of eight beer samples, each stored under three conditions: fresh, cold temperature, and room temperature. Each sample was injected in triplicate for UHPLC–MS acquisition, totaling 72 data files. Unlike a targeted metabolite experiment, the nontargeted focus of the study meant that every mass observed by the instrument served as its own data point. In this case, 7000 distinct molecular features were detected. Thus, the first step in data analysis involved peak picking and normalization in each individual data file, followed by retention-time alignment across all of the data files. The final output of these steps was a data matrix of molecular features defined by retention time, accurate mass, and normalized intensity in each data file.
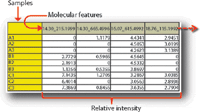
Figure 6: Example data matrix output from a nontargeted metabolite-profiling experiment. Samples A1âA3 represent triplicate analytical injections of the same sample. Molecular features are described by retention time (min)_accurate mass. Relative intensity values represent postnormalization values.
Figure 6 shows an example data matrix that can be used for various downstream statistical analyses. To determine molecular features that were potential "markers" of beer storage, we performed a simple analysis of variance (ANOVA) with "storage condition" and "tank" (to account for variables during brewing) as main effects. Additionally, we performed a principal component analysis (PCA), to assess global (multivariate) separations within the study. The results revealed 452 molecular features associated with storage conditions. PCA also revealed significant grouping of the data according to storage conditions (Figure 7). Taken together, these results indicate that each storage condition is described by a unique nonvolatile metabolite profile, and that beer stored in a cold-temperature environment was more similar to fresh beer than beer stored at room temperature.
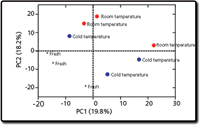
Figure 7: Principal component analysis (PCA) illustrating separation between fresh beer, beer stored cold, and beer stored at room temperature. Adapted from Heubeger and colleagues (10).
The detection of molecular isotopes, adduct formation, neutral losses, and in-source fragmentation lead to various molecular features that represent the same metabolite. After manual data interpretation, we determined that 16 unique metabolites varied significantly according to beer storage conditions. Metabolite annotation was performed using the following process.
- Accurate mass measurements were searched against the publicly available metabolite databases Metlin, Mass Bank, and National Institute of Standards and Technology (NIST).
- Using accurate-mass and isotopic distribution, molecular formulas were predicted and applied as a filter to the database search results.
- When possible, subsequent experimentation was performed to acquire MS-MS spectra for the significant molecular feature as well as for an authentic standard of the putative metabolite.
Identification confidence was assigned based on metabolomics standards initiative recommendations (11). Level 1 indicates confident molecular identification based on orthogonal, analytical parameters (accurate mass, retention time, and MS-MS fragmentation) relative to an authentic compound. Level 2 indicates a putative identification based on physicochemical properties and spectral similarity with literature or spectral libraries. Level 3 indicates putative identification of a compound class based on physicochemical properties or spectral similarity, and level 4 indicates an unknown compound.
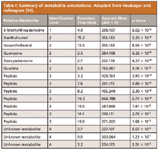
Table I: Summary of metabolite annotations. Adapted from Heubeger and colleagues (10).
The results of this annotation process appear in Table I. A comparison of the metabolite trends across storage conditions for a variety of the annotated metabolites illustrates the linear response of 5-methylthioadenosine (5-MTA) and highlights the potential of this metabolite as a molecular marker of beer aging. A commercial standard was subsequently used to validate the identification of this compound (Figure 8). This study represents the first observation of 5-MTA in beer, and the mechanism of its formation is unknown. Follow-up studies (data not shown) detected 5-MTA in other brews and observed similar trends with various aging conditions. The utility of 5-MTA as a marker of aging can be exploited to develop accelerated beer-aging models. Doing so would enable faster testing of new processes and ingredients to prolong flavor stability and shelf life.
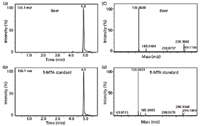
Figure 8: Level 1 identification of 5-MTA. Extracted ion chromatograms of 136.1 m/z from analysis of (a) beer and (b) a 5-MTA authentic standard. MS-MS fragmentation spectra of 136.1 m/z from the analysis of (c) beer and (d) a 5-MTA authentic standard. Adapted from Heubeger and colleagues (10).
Conclusions
Nontargeted metabolite profiling is a powerful approach that can lead to novel discoveries. While much attention is placed on the use of this approach for the study of human disease, the analytical approach does not depend on sample type and can be easily applied in many scientific fields. The example presented here illustrates the successful use of this approach in a food chemistry application.
Acknowledgment
We thank Corey Broeckling for contributions to the nontargeted metabolite workflow, and we acknowledge the contributions of Matthew R. Lewis, Lauren Salazar, and Peter Bouchaert to the beer study.
References
(1) C.D. Broeckling, A. Heuberger, and J.R. Prenni, J. Visualized Exp., in press (2013).
(2) W.B. Dunn et al., Chem. Soc. Rev. 40(1), 387–426 (2011).
(3) A.I. Nesvizhskii, O. Vitek, and R. Aebersold, Nat. Methods 4(10), 787–97 (2007).
(4) C.A. Smith et al., Ther. Drug Monit. 27(6), 747–751 (2005).
(5) H. Horai et al., J. Mass Spectrom. 45(7), 703–714 (2010).
(6) G. Theodoridis, H.G. Gika, and I.D. Wilson, Mass Spectrom. Rev. 30(5), 884–906 (2011).
(7) D.S. Wishart, Bioanalysis 3(15), 1769–1782 (2011).
(8) M.D. Wallenstein et al., Soil Biol. Biochem. 42(3), 484–490 (2010).
(9) D.S. Wishart, Trends Food Sci. Technol. 19(9), 482–493 (2008).
(10) A.L. Heuberger et al., Food Chem. 135(3), 1284–9 (2012).
(11) L.W. Sumner et al., Metabolomics 3(3), 211–221 (2007).
Jessica Prenni is Director of the Proteomics and Metabolomics Facility and Associate Professor in the Department of Biochemistry and Molecular Biology at Colorado State University in Fort Collins, Colorado. She has more than 11 years of experience in biological mass spectrometry and has authored over 36 peer-reviewed manuscripts in this area. The primary focus of the research in her group is the development of new approaches to improve the process of metabolite annotation in nontargeted metabolite profiling experiments by mass spectrometry.

Jessica Prenni
Adam Heuberger is a postdoctoral scientist in the Proteomics and Metabolomics Facility at Colorado State University. His research focuses on the application of both targeted and nontargeted metabolite profiling for the analysis of various sample types, including beer, malt, barley, and beans.

Adam Heuberger
Dana Sedin is the manager of the Sensory and Analytical laboratories at New Belgium Brewing Company in Fort Collins, Colorado. He has more than 12 years of experience as a brewing scientist.

Dana Sedin
Kate Yu "MS — The Practical Art" Editor Kate Yu joined Waters in Milford, Massachusetts, in 1998. She has a wealth of experience in applying LC–MS technologies to various application fields such as metabolite identification, metabolomics, quantitative bioanalysis, natural products, and environmental applications. Direct correspondence about this column to lcgcedit@lcgcmag.com

Kate Yu

Analytical Challenges in Measuring Migration from Food Contact Materials
November 2nd 2015Food contact materials contain low molecular weight additives and processing aids which can migrate into foods leading to trace levels of contamination. Food safety is ensured through regulations, comprising compositional controls and migration limits, which present a significant analytical challenge to the food industry to ensure compliance and demonstrate due diligence. Of the various analytical approaches, LC-MS/MS has proved to be an essential tool in monitoring migration of target compounds into foods, and more sophisticated approaches such as LC-high resolution MS (Orbitrap) are being increasingly used for untargeted analysis to monitor non-intentionally added substances. This podcast will provide an overview to this area, illustrated with various applications showing current approaches being employed.