Mass Spectrometry for Natural Products Research: Challenges, Pitfalls, and Opportunities
LCGC North America
Correlating biological activity with chemical composition is the Holy Grail of natural product chemists. Is mass spectrometry the right tool for this challenge?
A common attitude in natural products research is that nuclear magnetic resonance (NMR) spectroscopy serves as a primary tool, whereas mass spectrometry (MS) is relegated to the task of providing the molecular formulas of pure compounds. Yet over the past several decades, we have witnessed astonishing growth in MS. Electrospray ionization has enabled the analysis of biological molecules previously deemed intractable, and instruments that offer astounding mass accuracy are becoming routinely available. Nonetheless, as applied to natural products research, MS is still fraught with challenges and pitfalls. Here is an account of strategies to conduct effective research despite these obstacles.
Among natural products chemists there is a joke that goes like this: Nuclear magnetic resonance (NMR) spectroscopy is like your mother; she knows what is good for you and tells you what you need to hear. Mass spectrometry (MS) is like your lover, willing to say whatever you want to hear whether it is true or not.
This joke reflects a common attitude in natural products research; NMR serves as a primary tool, whereas MS is relegated to the task of providing the molecular formulas of pure compounds. Yet over the past several decades, we have witnessed astonishing growth in MS. Electrospray ionization (ESI) has enabled the analysis of biological molecules previously deemed intractable, and instruments that offer astounding mass accuracy are becoming routinely available. Nonetheless, as applied to natural products research, MS is still fraught with challenges and pitfalls. What follows is an account of the strategies that my laboratory (and those of colleagues and collaborators) espouse to conduct effective research despite these obstacles. In this column, I have tried to paint an honest picture of what we actually do (and don't do) in the laboratory rather than what is theoretically possible. As such, this account reflects my own perspective and opinions. By no means do I present it as the only (or last) word on the topic.
MS for Structure Elucidation
Natural products research is primarily concerned with identifying useful compounds from natural sources, including plants, fungi, bacteria, and marine organisms. These organisms share the common characteristic of being complex mixtures of thousands of structurally diverse molecules present at varying abundance (1). (Note that reference 1 describes a typical plant extract mixture in which there are estimated to be 920 compounds theoretically detectable by a given liquid chromatography with ultraviolet absorbance detection [LC–UV] method. These represent only a subset of the compounds present in the mixture.) Natural products chemists seek to unravel this complexity and distill it to the key active elements that contribute to a desired effect. Thus, we isolate anti-inflammatory compounds from marine sponges, insecticidal compounds from fungi, or antimicrobial compounds from traditional plant-derived medicines.
Like many scientists educated in the 1990s, I can thank Sean Connery — or, more precisely, his character in the film "Medicine Man" — for my introduction to natural products research. Those acquainted with this classic may recall a scene in which a cancer-curing natural product mixture is injected into a portable gas chromatography–mass spectrometry (GC–MS) system, which, having been transported by canoe, miraculously operates in the midst of the jungle on generator-provided power. Within seconds, the structure, including stereochemistry, of a new molecule responsible for the biological activity of this mixture appears on a blinking LED screen. Fantastic as it seems even by 2013 standards, this scenario could represent the holy grail of natural products research. Such an instrument — portable and able to elucidate the structure of components in a mixture without the need for user intervention or pure standards — would revolutionize our field, not to mention all of chemistry and biology. Regrettably, analytical equipment of such awesome capability does not currently exist. The tool that comes closest, however, is the mass spectrometer.
Advantages and Disadvantages of MS as a Tool for Solving Structures
Molecular mass is an intrinsic quality of any analyte undergoing measurement that, by means of MS, we can easily calculate a priori. Despite this faculty, we mass spectrometrists are not particularly good at de novo structure elucidation for unknown small molecules. MS data provide an excellent complement to NMR data for solving unknown structures and are useful for rapidly searching a database to determine whether a compound was previously identified. Those benefits notwithstanding, NMR is the far more effective technique for solving unknown structures, provided sufficient quantities of purified material are available. It is hardly surprising, then, that in natural products research, where identifying compounds is the most critical goal, scientists expert in isolation and NMR structure elucidation dominate. Unfortunately, this reliance on NMR has encouraged a bias toward structurally interesting and easily isolable compounds. Such compounds often become the major focus, even at the expense of those that are more biologically interesting. Also, as we shall later see, combination effects (synergy) may be overlooked in the isolation process. Finally, to achieve pure samples for NMR analysis requires an intense isolation effort, one often wasted when applied to known compounds, particularly commercially available ones. MS can enable identification without isolation, so it offers the potential to resolve many of these issues. Still, limitations loom, and they must be addressed before we can consider MS an optimal technique for structure elucidation.
How GC–MS Is Applied for Unknown Identification
GC–MS instruments are arguably the best mass spectrometers for identifying unknown small molecules. Their effectiveness stems from compatibility with electron ionization (EI), which generates highly consistent and searchable fragmentation spectra. By searching an unknown against spectral libraries, one can generate candidate structures in seconds. For example, the National Institute of Standards and Technology (NIST) standards database contains over 200,000 EI spectra. Despite the availability of such excellent databases, however, the limitations of EI hinder our ability to identify unknown compounds. EI's harshness often makes detecting the molecular ion difficult, and the ability to identify compounds is limited by information previously entered into the database. Also, EI spectra often do not enable isomers to be distinguished. Therefore, identifications made on the basis of mass spectral databases are always tentative and must be confirmed with an orthogonal technique like NMR. Even so, we can gain much by using MS to obtain a tentative structure before beginning the isolation process. The MS result often facilitates a decision about whether isolation is worthwhile (on the basis of a compound's novelty) and simplifies isolation and structure elucidation.
How Liquid Chromatography–Mass Spectrometry Is Applied for Unknown Identification
Most biologically interesting molecules, including the majority of natural products, are nonvolatile and, therefore, unsuitable for direct analysis by GC–MS. Even more problematic, most biologically relevant matrices (including many natural product extracts) are also nonvolatile. ESI-MS can ionize nonvolatile species, enabling direct coupling between LC and MS. Its application for this purpose has become so commonplace that the term LC–MS typically implies the use of ESI. Before the advent of electrospray, analysts routinely spent much time and effort extracting and derivatizing biological samples to render them suitable for GC–MS analysis. Nowadays, this process is commonly skipped in favor of LC–MS, for which sample preparation is much simpler.
The approach for identifying unknowns using LC–MS is fundamentally different than that employed for GC–MS. ESI yields molecular or pseudomolecular ions without appreciable fragmentation for many analytes. Thus, coupling ESI to an instrument with accurate-mass capabilities (such as an Orbitrap [Thermo Scientific] or quadrupole time-of-flight [QTOF] mass spectrometer) enables rapid determination of the molecular formula of interest. A caveat is that in-source fragmentation (for example, the loss of water) and adduct formation (for example, with sodium or acetate ions) may complicate the process of identifying the molecular ion. Isotope ratios inherent in the MS data can help confirm correct formula assignment, and various software packages for deconvoluting MS and LC–MS data, such as IntelliXtract (ACD/Labs), ExactFinder (Thermo Scientific), and Apex (Sierra Analytics), can identify likely clusters and fragments according to characteristic mass differences. It is also possible (and highly advisable) to manually inspect mass-spectral data for characteristic mass differences among peaks. For example, formation of a sodium cluster will result in a 22 Da mass difference between the mass-to-charge ratio (m/z) of the [M + H]+ ion and the [M + Na]+ ion, and neutral loss of water will result in a difference of 18 Da. Assignments of molecular formulas based on MS data alone, however, should always be viewed with caution. A common mistake is to search structural databases using the molecular formula of a fragment or cluster ion, find nothing, and then incorrectly conclude that the structure was not previously reported.
Assuming that formulas are correctly assigned, they can be searched against those of known compounds as a first stage in identification. Useful though this approach is, it doesn't solve the de novo structure-elucidation quandary. Many molecules share the same molecular formula and, consequently, are indistinguishable by accurate mass and isotope patterns alone. For example, literally hundreds of isobaric (same mass) flavonoids have been identified from plants, and searching a flavonoid molecular formula in the Dictionary of Natural Products (CRC CHEMnetBASE database) or SciFinder (CAS, a division of the American Chemical Society) can yield countless hits. The structure of the true analyte may or may not be included among these, depending on whether it was previously reported and uploaded to the relevant database. To assign the correct structure from among isobaric candidates, we need orthogonal information. NMR data are best for this purpose, assuming the availability of sufficient purified analyte. Where an analyte has not been purified, applying tandem MS (MS-MS) measurements is the next-best tool for structure confirmation. The term "MS-MS" refers to a two-stage MS experiment whereby an ion of interest (the precursor ion) is selected in the first stage and then fragmented. The masses of the fragments are then measured, constituting the second stage. Multiple methods of generating fragment ions for MS-MS experiments exist. Among the most common is collisionally induced dissociation (CID), wherein the precursor ion collides with gas molecules and thus fragments. When the fragments are formed from high-energy collisions, the technique is called higher-energy collisional dissociation (HCD). Many mass analyzers enable the generation of MS-MS data, including triple-quadrupoles, ion-trap, and QTOF instruments. However, the relative abundance and presence or absence of particular fragments can vary greatly by instrument. Even for a single instrument, such variation can arise from changing parameter settings. Thus, it is currently common practice to use MS-MS spectra primarily as a tool to compare standards with unknowns under the same conditions on the same instrument. In this way, structural assignments can be made by matching retention time and MS-MS spectra. Unfortunately, the availability of the required standards is extremely limited, which restricts the applicability of this approach.
Difficulty in assigning structure based on LC–MS data is a critical barrier today in the field of natural products research. This barrier could be surmounted if a comprehensive and searchable database of MS-MS spectra analogous to that available for GC–MS data existed. Of the databases that seek to fill this need, none are perfect for natural products applications. Currently, the best databases for searching natural product molecular formulas are the Dictionary of Natural Products, AntiMarin (University of Canterbury, New Zealand), and SciFinder. None of these are open-access or completely comprehensive, nor do they provide searchable MS-MS spectra. Open-access, searchable databases for small molecules do exist — for example MassBank (MassBank Project, Keio University, Tsuruoka City, Yamagata, Japan) and ChemSpider (Royal Society of Chemistry). None, however, are tailored specifically to natural products. Admittedly, generating a comprehensive database of natural product MS-MS spectra would be a far-from-trivial accomplishment. Doing so would require empirical measurements made from standards of all molecules of interest. Furthermore, such a database, if compiled, would also need to address issues of limited reproducibility among different instruments. Only through the combined efforts of many researchers could such a natural product database be compiled. Moreover, those researchers would need access to diverse natural product standards. Finally, they would require expertise in analyzing data presented in numerous software formats and in collecting and interpreting those data on multiple MS platforms.
It is tempting to propose that the requirement of empirically measured MS-MS spectra could be circumvented by generating predicted spectra from known molecular structures. Indeed, excellent databases of predicted fragment spectra are available for peptides, and the field of proteomics would not exist in its current, advanced form without these databases. Useful, predicted MS-MS spectra for peptides can be generated because when these compounds are subjected to collisionally induced dissociation, they fragment consistently. Unfortunately, the structural diversity of natural product secondary metabolites makes developing rules to predict their MS-MS spectra difficult. Current rule-based software packages for this purpose (for example, Mass Frontier [Thermo Scientific], ACD/MS Fragmenter [ACD/Labs], and MassFragment [Waters]) generate hundreds of predicted fragments with no indication as to which will be observed experimentally. Thus, these tools are truly useful only for assigning observed fragment structures. A comprehensive empirical database of high-resolution mass spectra and MS-MS spectra for diverse structural classes of natural products might enable improvements in the currently available fragmentation predictors.
Selectivity in LC–MS Analysis of Natural Products
It is the selectivity of MS — its tendency to generate radically different responses for analytes with different structures — that led to its playful characterization among natural products chemists as the misleading mistress. Selectivity in MS analyses is primarily dictated by the type of ionization source used. No truly universal ionization technique exists for LC–MS. ESI, the most commonly used ionization technique, is selective for analytes that contain an inherent positive charge or that can be charged by protonation, deprotonation, or adduct formation. Among charged analytes, an additional layer of selectivity is introduced by the analyte's nonpolar character, with nonpolar, chargeable analytes yielding the highest response (2). Although many natural products are ionizable by ESI, they can vary widely in responsiveness. Several common contaminants, such as polymers and surfactants, yield high signals and can suppress the response of other analytes of interest. LC–MS analysts must, therefore, resist the tempting assumption that the largest peaks in the total ion current (TIC) chromatogram represent the most important — or even the most abundant — compounds in the sample.
Despite issues of selectivity, it is possible to compare the relative abundance of the same compound in different samples on the basis of peak area in LC–MS chromatograms. In many applications of natural products chemistry — for example, the evaluation of purity — being able to compare the relative abundance of different classes of compounds in a single sample or multiple samples would be desirable. Differences in ionization efficiency prohibit such comparisons on the basis of data obtained by electrospray ionization. Instead, an orthogonal detection technique, where response is more closely correlated with analyte abundance, must be employed. Two popular techniques for this purpose are evaporative light-scattering detection (ELSD) and charged-aerosol detection (CAD). These detection methods do not replace LC–MS. They are far less sensitive and do not provide structural information. Nevertheless, comparison of ELSD or CAD chromatograms with those obtained with LC–MS can help discern which peaks correspond to the most abundant compounds in a sample.
Although most LC–MS separations employ ESI for ionization, other ionization techniques are sometimes used to provide complementary information or detect small molecules not ionizable with ESI. The most common of these are atmospheric pressure photoionization (APPI) (3) and atmospheric pressure chemical ionization (APCI). APCI is harsher than ESI and may ionize polar compounds that lack acidic or basic functional groups. APPI can be applied for the analysis of some nonpolar species that are not amenable to ESI analysis, and studies suggest that this technique is somewhat more universal than ESI for small drug-like molecules (4,5). On the other hand, APPI and APCI source development has been less a focus of instrument companies than ESI sources, which are typically better designed. APPI also suffers from practical limitations, including the need to introduce dopant and the expense of replacing source bulbs. APCI can often achieve better linear dynamic ranges than those possible with ESI for some analytes, but APCI typically yields slightly higher limits of detection (6). Practically, the need to switch sources and collect copious quantities of data often limits the ability to operate using multiple ionization modes. For many applications, collection of data in the positive and negative ionization modes using LC–MS with ESI is sufficient. However, the possibility of missing active compounds because of the selectivity of this technique should always be considered. Finally, even with access to APCI, ESI, and APPI, ionization is impossible for some natural product molecules. A need remains for better methods to ionize small molecules that prove intractable using existing techniques.
MS for Quantitative Analysis of Mixture Components
Natural products chemistry often involves quantitative analysis. Such analysis is necessary to verify the solubility or extractability of natural product molecules in different solvents, compare levels of bioactive compounds in different raw materials, or measure toxins or contaminants. MS is particularly useful for such applications. It can detect compounds present in minute abundance (sub-parts-per-billion levels) and resolve molecules on the basis of their m/z values. Indeed, because of their exceptional sensitivity and selectivity, triple-quadrupole mass spectrometers operating in the selected reaction monitoring (SRM) mode have long been considered the gold standard for the quantitative analysis of trace mixture components. Recently developed high-resolution MS platforms, such as the LTQ Orbitrap and Q Exactive systems (both from Thermo Scientific), enable quantitation based on accurate-mass measurements of the molecular ion. Such quantitation now rivals the selectivity of triple-quadrupole instruments (7,8).
Absolute quantitation with MS requires pure standards. Although some standards can be obtained commercially (for example, from SigmaAldrich or ChromaDex), in many cases the compounds of interest are unavailable. Options are to rely on relative, rather than absolute, quantitation or to synthesize or isolate the standard of interest. Obtaining natural product standards that are sufficiently pure to accomplish absolute quantitation is a major challenge, one that applies even with commercial standards. It is common practice to report the purity of these standards according to their response in LC–UV chromatograms. Yet this approach detects only contaminants with UV chromophores. Quantitative NMR can circumvent this problem (9) but is not yet widely adopted as a method to evaluate purity. Thus, the accuracy of percent purity claims for commercial standards is questionable.
It is common practice when performing quantitative analysis via MS to distinguish coeluted compounds by plotting selected ion chromatograms. Here the phrase "selected ion chromatogram" refers to a plot of ion current versus time for the m/z value of the compound of interest. Such selected ion chromatograms can be used to plot calibration curves even for compounds with identical retention times. Chemists trained to perform quantitative analysis using LC–UV are driven absolutely mad by this practice. They invariably cite the concern of ion suppression (otherwise referred to as matrix interference) — that one ion may influence the ionization of another, thereby skewing the quantitative results.
If the goal of a quantitative analysis is relative quantitation, and if the matrix is consistent among samples, ion suppression is not a major concern. However, matrix suppression can seriously undermine the accuracy of measurements seeking to determine absolute concentration. The chromatographer's solution to this problem is to separate all components of the mixture with baseline resolution and quantify by LC–UV. However, for most biologically relevant complex mixtures, including complex natural product extracts, the physical limitations on chromatographic resolution (10) make doing so impossible in a single stage of separation. In fact, what may in LC–UV appear to be baseline separation is merely baseline resolution of all components detectable by the UV detector, a fact revealed when LC–UV and LC–MS data are compared for the same sample (Figure 1).
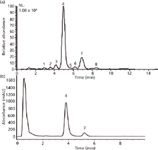
Figure 1: Comparison of (a) LCâMS and (b) LCâUV chromatograms for the same botanical extract. The peaks in the chromatogram represent a series of alkylamides extracted from the plant Spilanthes acmella (11). The chromatogram in (a) is a base peak chromatogram normalized to a total ion count of 1.08 Ã 109. Only two of the eight compounds detectable by LCâMS can be detected with LCâUV.
When mixture components cannot be fully resolved chromatographically from others with similar absorbance, or when they are present below the limit of quantitation with a UV detector, quantitation by LC–MS is often a valid alternative to quantitation by LC–UV. For such analyses, the extent of matrix interference must be evaluated by comparing the signal (peak area) for standards in solvent versus matrix. This approach is necessary because matrix interference can be caused by compounds that the mass spectrometer cannot detect, such as salts that wash off the column early in the separation process. Thus, matrix effects can occur even when the mass spectrum appears to contain only one compound. A number of strategies can be used to address matrix effects, including standard addition and matrix matching. A recent report by Kruve and Leito (12) nicely demonstrates strengths and limitations of these and other approaches.
However tempting (particularly for analytical chemists) is the quest for absolute quantitation, it may not always be necessary. When standards are not available, it is possible to compare relative peak areas for a particular sample component to draw useful conclusions. When making such comparisons, it is important to be aware of biases that systematic fluctuations in instrument response may introduce. A common source of such fluctuations is fouling of ion optics over time, which may cause the signal intensity to decrease slowly during the analysis. Including control standards at the beginning, middle and end of a run can help identify this problem. Changes in ion transmission that result from tuning or cleaning can also cause the absolute instrument response to shift (consistently higher or consistently lower) between runs. For this reason, collecting all data for a relative quantitation experiment in a single analysis is advisable. If doing so is impractical, data from the same sample or standard analyzed (with replicates) in each run should be compared to ensure that the response did not drift (beyond the tolerance of random errors) between analyses. If such drift does occur, adjusting for it is extremely difficult. Theoretically, comparing the peak area of each analyte to that of an internal standard can correct for differences in instrument response between runs. Practically, however, this approach rarely seems to work, perhaps because signal drift does not occur consistently across the chromatogram.
The Holy Grail of Natural Products Chemistry: Correlating Biological Activity with Chemical Composition
The truly interesting questions in natural products chemistry occur at the intersection of chemistry and biology. These questions seek information about how natural products can be used to treat diseases, eliminate pests, produce greener technologies, or generally benefit the planet or humankind. To address such questions, it is not enough to simply determine what is in a sample. Rather, we want to answer the question "what are the bioactive components of this sample?" The following paragraphs describe some of the traditional methods for addressing this question, their limitations, and potential areas in which MS can help resolve those limitations. (Here I should note that biologists would pose a different question as the Holy Grail of natural-products research: What is the mechanism of action for bioactive natural products? MS certainly can contribute something toward investigating mechanistic questions as well, but that topic extends beyond the scope of this column installment.)
Strengths and Limitations of Bioactivity-Guided Fractionation for Active Compound Identification
Natural products researchers typically rely on bioactivity-guided fractionation to identify the active compounds in a mixture. To apply this technique, a series of natural product extracts (from plants, fungi, bacteria, marine organisms, or other natural sources) are screened against a desired biological assay (cytotoxicity, antimicrobial, insecticidal, and so forth) After an extract is identified as a good lead — one that evidences the desired biological activity — it is separated, usually by liquid–liquid partitioning or column chromatography. The resultant fractions are then tested using the same biological assay, and the active fractions undergo further purification. This process is repeated iteratively until compounds of sufficient purity for structure elucidation and more in-depth evaluation of activity are isolated.
It is highly undesirable, wasting both time and resources, for the bioactivity-guided fractionation process to result in known compounds with known activity. Natural products chemists therefore employ various "dereplication" strategies to prevent the reisolation of known compounds. The ideal dereplication strategy would rapidly identify all known compounds in a mixture without any prior purification. Both MS (13) and NMR (14) strategies have been pursued toward this goal. Dereplication by MS is currently seriously limited by the aforementioned lack of searchable databases of LC–MS data. To circumvent this problem, many laboratories construct in-house databases to enable the dereplication of compounds commonly encountered in their samples of interest. For example, my collaborator Nicholas Oberlies at the University of North Carolina Greensboro developed a 170-compound library with MS, MS-MS, UV, and retention-time data that can identify fungal secondary metabolites previously isolated by his laboratory (15). Using this database, it is possible to rule out ~50% of the new fungal samples the Oberlies laboratory extracts because of the dominance of compounds his laboratory has already isolated. Similar in-house databases in commercial and academic research laboratories around the world hold a treasure trove of information that if combined into a single natural products database, would be a formidable tool indeed.
A major challenge associated with bioactivity-guided fractionation is the inherent assumption that the bioactivity of a mixture can be distilled to a single compound (or series of compounds). Certainly, historical precedent for such an assumption exists. Key examples include taxol, from yew bark, and camptothecin, from the Chinese tree Camptotheca acuminata (16). Both were isolated by Wall and Wani (17)using bioactivity-guided fractionation, and both became highly effective cancer drugs when used in isolation (that is, without any other components of the mixture). In many cases, however, particularly those involving botanical extracts, all or part of the activity is lost in the isolation process. Moreover, this loss may not always be readily apparent. Because mixture components are purified and tested for activity at increasingly higher concentrations with each successive stage of isolation, a perceived improvement in activity upon purification may actually be attributable to an increase in concentration. To determine whether a particular isolated compound represents the entire activity of a mixture, it is useful at the end of a bioactivity-guided fractionation experiment to compare the activity of the pure component to that of the original mixture at identical concentrations. Such comparisons require the concentration of the bioactive compound to be determined in the original mixture. Fortunately, the bioactivity-guided fractionation process itself produces the pure standard needed to measure this concentration.
If, as is often the case, the mixture displays better activity than the pure compounds, the explanation is typically that some sort of synergy is involved in the activity of the mixture. Here synergy is defined as a scenario in which the whole is greater than the sum of its parts. The underlying mechanisms for synergy in complex mixtures, which have been reviewed elsewhere, may include several compounds interacting at the same receptor, multiple compounds targeting different biological receptors, or situations where the solubility of one compound is improved by the presence of another. This last case is surprisingly common with in vitro assays of natural products. Many natural products chemists have encountered the maddening experience of beginning with a perfectly soluble (and bioactive) mixture, and after multiple painstaking steps of isolation ending up with a pure compound that is about as soluble (and bioactive) as brick dust.
Recombining fractions from an active extract is sometimes proposed as a way to identify synergists. It is not practical, however, to test the vast number of fractions generated by a bioassay-guided fractionation experiment in combination. For example, if such an experiment generates 10 fractions in the first stage of separation, those fractions, taken two at a time, can be combined in 45 different ways: n!/(k!(n – 2)!, where n = 10 and k = 2. To accomplish combination assays of these fractions over a relevant concentration range would require an estimated 9000 assays. If at least three stages of fractionation are needed, the number of assays required increases exponentially to more than 1,000,000. Even this large figure ignores the possibility that more than two fractions may be required to achieve synergy.
Why Isolate at All?
Given the limitations of bioassay-guided fractionation, it might seem advisable to perform all biological assays on mixtures, without isolation. Unfortunately, correlating the presence of components in a complex mixture with its biological activity is difficult. A too-common scenario in the botanical medicine literature involves testing a mixture for some biological effect, analyzing it for the presence of known "marker" compounds, and then surmising (hoping?) that these compounds produce that effect. The choice of marker compounds is often made for practical reasons, such as the availability of standards or the ease with which they are detected. Testing for the presence of these marker compounds is prudent, to authenticate the source material, but does not of itself establish a link between activity and chemical composition. Sufficient statistical power to sort the components responsible for activity requires the number of measurements of biological activity to be at least as great as the number of compounds present, which is impossible if activity is tested for a single mixture. Sufficient statistical power theoretically can be achieved if multiple mixtures containing a range of concentrations of bioactive molecules are tested. Carrying out such testing, however, requires some fractionation, so we find ourselves back where we started.
Synergy-Guided Fractionation to Address the "Kobayashi Maru" of Natural Products
The preceding section describes what appears to be a no-win situation, a "Kobayashi Maru" of natural products. (For the enlightenment of those who are not acquainted with Star Trek, the "Kobayashi Maru" is a test in the fictional Star Trek universe for which there is no solution.) To investigate the activity of mixtures makes identifying the biologically active components difficult, but isolating the components from the mixture often results in loss of activity because of the inability to account for synergy. Our laboratory has worked to develop strategies to address this dilemma. Currently, we are conducting several studies for which the goal is to identify the array of compounds responsible for biological activity of botanical medicines. To accomplish this goal, we developed a modification of the bioactivity-guided fractionation approach that we refer to as "synergy-guided fractionation" (18) (Figure 2). This approach is similar to bioactivity-guided fractionation, except that fractions are tested in a synergy assay where they are combined with the original crude extract or some isolated component thereof. The synergy-guided fractionation approach solves the dilemma of generating an exponentially increasing number of combinations. It also ensures that all compounds of interest are present in any given biological assay. In analytical chemistry terms, such an experiment is essentially one of standard addition, but it is performed with a biological rather than a chemical endpoint.
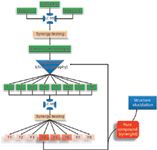
Figure 2: Schematic of synergy-directed fractionation. A series of extracts is profiled using LCâMS to identify known compounds (if possible) and is subjected to synergy assays. Extracts that demonstrate synergistic effects are subjected to separation, and the fractions are then tested again to identify which fractions contain synergists. This process is repeated iteratively until a pure compound is isolated in sufficient quantity for structure elucidation.
Our first case study of the synergy-directed fractionation approach involved applying it to identify bioactive flavonoids from the botanical medicine goldenseal (Hydrastis canadensis) (Figure 3) (18). These flavonoids were shown to significantly enhance the antimicrobial activity of the known alkaloid berberine, also a constituent of H. canadensis. We demonstrated that the ability of the flavonoids to act as efflux pump inhibitors caused this enhancement. Importantly, the flavonoids were inactive as antimicrobial agents alone, and a traditional bioactivity-guided fractionation approach would, therefore, have missed them. The synergy assays that led to the isolation of flavonoids from H. canadensis involved combining extract fractions with purified berberine. We are currently engaged in more detailed studies to identify additional synergists by testing in combination with the crude H. canadensis extract.

Figure 3: Synergy-directed fractionation of the medicinal plant goldenseal (Hydrastis canadensis) yielded the flavonoids sideroxylin (1), 8-desmethyl-sideroxylin (2), and 6-desmethyl-sideroxylin (3). These compounds enhance the antimicrobial activity of the alkaloid berberine (4) via efflux inhibition.
A Role for MS in Bioactivity-Guided and Synergy-Guided Fractionation?
Bioassay-guided fractionation and synergy-directed fractionation are time-consuming processes inherently biased toward isolable compounds. It would be very desirable to develop approaches to rapidly obtain a more comprehensive understanding of the relationship between the composition and biological activity of natural product mixtures. Toward this end, we have used MS to track compounds of interest throughout the isolation process by their measured m/z values and retention times. This practice has proven useful for verifying whether activity corresponds with compounds already reported as constituents of botanical interest (so far, generally, it has not). In addition, we have attempted, albeit with limited success, to use untargeted metabolomics to determine which compounds are unique to active fractions and to then focus our isolation efforts on those compounds. Theoretically, an advantage of this approach is that mass-guided fractionation (isolation focused on a particular ion) is far more efficient than bioassay-guided fractionation (isolation based on a particular biological activity). Yet a number of practical challenges prevent this approach from being entirely effective. First, for complex botanical extracts, a large number of fractions are needed to resolve mixture components sufficiently to distinguish bioactive from inactive compounds. It is difficult to identify a biological assay that is inexpensive, robust, and efficient enough for this purpose. Second, fractionation may again cause synergistic effects to be overlooked. Finally, the possibility remains that the most biologically interesting molecules in a given set of extract fractions are either undetectable, because of the selectivity of the mass spectrometer, or masked by other mixture components. The undetectable nature of such molecules renders fractionation based only on mass somewhat unnerving.
For all these reasons, we have yet to significantly improve the bioactivity-directed fractionation process by involving MS (beyond the initial dereplication step). Nonetheless, we expect that, with improvements in methodology, this will eventually become possible. These improvements may include the development of better databases for natural product identification, improved software methods for correlating biological and MS data, and more creative and robust biological assays.
The Future
Currently, many natural products chemists still ignore MS in the isolation process and instead employ NMR data to facilitate isolation of as many unique (and structurally interesting) compounds as possible. Given all of the challenges and limitations addressed here, this rejection of MS is perhaps unsurprising. It is clear that the mass spectrometer portrayed in "Medicine Man," one that can simultaneously solve natural product structures and identify those that are biologically active, does not yet exist. The development of such a mythical instrument will demand continued advances in the ion source universality, instrument sensitivity, dynamic range, resolving power, and — most importantly — software and database capabilities. Such accomplishments will require sustained collaborative efforts involving (but not limited to) analytical chemists, natural products chemists, instrument and software developers, and biologists. Nonetheless, as I gauge how far our field has progressed in recent decades, I am convinced that these advances are indeed possible, and that the future of MS as a tool for natural products research is unquestionably bright.
References
(1) C.G. Enke and L.J. Nagels, Anal. Chem. 83(7), 2539–2546 (2011).
(2) N.B. Cech and C.G. Enke, Rev. Mass Spectrom. 20(6), 362–287 (2002).
(3) D.B. Robb, T.R. Covey, and A.P. Bruins, Anal. Chem. 72(15), 3653–3659 (2000).
(4) Y. Cai, D. Kingery, O. McConnell, and A.C. Bach, Rapid. Commun. Mass Spectrom. 19(12), 1717–1724 (2005).
(5) D.B. Robb and M.W. Blades, Anal. Chim. Acta 627(1), 34–49 (2008).
(6) L.C. Herrera, J.S. Grossert, and L. Ramaley, J. Am. Soc. Mass Spectrom. 19(12), 1926–1941 (2008).
(7) A. Kaufmann, P. Butcher, K. Maden, S. Walker, and M. Widmer, Anal. Chim. Acta 673(1), 60–72 (2010).
(8) A. Kaufmann, Anal. Bioanal. Chem. 403(5), 1233–1249 (2012).
(9) M. Weber, C. Hellriegel, A. Ruck, R. Sauermoser, and J. Wuthrich, Accred. Qual. Assur. 18, 91–98 (2013).
(10) J.M. Davis and J.C. Giddings, Anal. Chem. 55(3), 418–424 (1983).
(11) S.S. Bae, B.M. Ehrmann, K.A. Ettefagh and N.B. Cech, Phytochem. Anal. 21(5), 438–443 (2010).
(12) A. Kruve and I. Leito, Anal. Methods 5, 3035–3044 (2013).
(13) K.F. Nielsen and J. Smedsgaard, J. Chrom. A 1002(1-2), 111–136 (2003).
(14) G. Lang et al., J. Nat. Prod. 71(9), 1595–1599 (2008).
(15) T. El-Elimat et al., J. Nat. Prod. in press (2013).
(16) W.-L. Lee, J.-Y. Shiau and L.-F. Shyur, Adv. Bot. Res. 62, 133–178 (2012).
(17) M.E. Wall and M.C. Wani, J. Ethnopharmacol. 51(1-3), 239–254 (1996).
(18) H.A. Junio et al., J. Nat. Prod. 74(7), 1621–1629 (2011).
Nadja B. Cech, PhD, earned her BS degree in chemistry from Southern Oregon University in 1997, and her PhD in Analytical Chemistry from the University of New Mexico in 2001. Her PhD training is in the area of mass spectrometry, and for the last 14 years she has worked to apply this expertise to solve challenging problems in natural products research. As a faculty member at the University of North Carolina Greensboro, Dr. Cech supervises a research group of 12 students and postdoctoral research associates. She is the recipient of the 2011 Journal of Natural Products Jack L. Beal Award, for a paper detailing approaches to study synergy in botanical medicines. Dr. Cech is funded by the National Institutes of Health on several projects that involve the identification of botanical products effective against inflammation or infection.

Nadja B. Cech, PhD
Kate Yu "MS — The Practical Art" Editor Kate Yu joined Waters in Milford, Massachusetts, in 1998. She has a wealth of experience in applying LC–MS technologies to various application fields such as metabolite identification, metabolomics, quantitative bioanalysis, natural products, and environmental applications. Direct correspondence about this column to lcgcedit@lcgcmag.com

Kate Yu

University of Rouen-Normandy Scientists Explore Eco-Friendly Sampling Approach for GC-HRMS
April 17th 2025Root exudates—substances secreted by living plant roots—are challenging to sample, as they are typically extracted using artificial devices and can vary widely in both quantity and composition across plant species.
Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.
Silvia Radenkovic on Building Connections in the Scientific Community
April 11th 2025In the second part of our conversation with Silvia Radenkovic, she shares insights into her involvement in scientific organizations and offers advice for young scientists looking to engage more in scientific organizations.