Trends in Biopharmaceutical Analysis: A Focus on Integrating Single-Cell Omics with Microfluidic Chips
Biopharmaceutical analysis is a rapidly evolving field that requires the development of new technologies and methods to keep pace with the increasing complexity of biologics. One of the most promising areas of research is the use of single-cell omics and microfluidic chips for the analysis of biopharmaceuticals. Single-cell omics has revolutionized our understanding of cellular heterogeneity, while microfluidic chips have enabled high-throughput analysis of single cells that provide an understanding of the complex biological network that complements the genomics and transcriptomics studies. This article will explore some of the emerging trends and technologies in biopharmaceutical analysis, with a particular focus on single-cell omics and microfluidic chips. We will also discuss the developments in ambient ionization mass spectrometry such as sub nanoampere ionization and the potential of low current ionization in studying cell-to-cell heterogeneity and its role in metabolomics.
Variations displayed by the cell populations in their DNA, RNA, proteins, and metabolites lead to significant heterogeneity and differences in cell states (1). An understanding of the cell-to-cell variations can help reveal the physiological and pathological processes such as disease progression (2). While traditional methods in cell analysis have overlooked cell-to-cell variations that contribute to drug resistance, the development of single-cell analysis methods has enabled remarkable progress in clinical and biomedical research particularly in drug development (3).
While the molecular copy numbers of nucleotides, proteins, and metabolites in single cells are low, the dynamic range is wide; therefore, highly sensitive single cell sample preparation methods are of great value for single cell analysis (4). In this regard, microfluidics has emerged as an important tool in single-cell analysis with the advantages of cost effectiveness, automation, miniaturization, and high throughput ability (5,6).
In recent years, there has been a growing interest in integrating single-cell omics with microfluidic chips for the analysis of biopharmaceuticals (7,8). This approach has several advantages over traditional methods of biopharmaceutical analysis, as it allows for a high degree of automation and allows for high-throughput studies due to its ability to perform multiple assays on a single chip. Minimum amounts of reagents are required, reducing assay cost, and improved sensitivity is achieved by using microchannels that provide a higher surface-area-to-volume ratio. The multiple unit microfluidic chip allows for the different processes of cell injection, culture, capture, lysis, and detection to be completed in a single microfluidic chip. In addition, multiple structures can also be included in a microfluidic chip to capture single cells. At a drug discovery level, microfluidics can help in identification, characterization, purification, and structure elucidation of chemical moieties (9). Drug screening can achieve high throughput, cost-effective, and rapid increase in the rate of sample analysis using microfluidics.
A microfluidic chip is mostly fabricated with elastomers, inorganic materials, and thermoplastics for either droplet microfluidics, microwell arrays, or hydrodynamic microfluidics (such as S-shaped microchannels). The commonly used fabrication materials include polydimethylsiloxane (PDMS), silicon, glass, quartz, polycarbonate, and polymethylacrylate (PMMA). The chip comprises of a reagent inlet, sample inlet, valves, microchannels, the drainage system, and the sensor part (10). Based on the flow of fluids, microfluidic chips can be classified into three main types: traps-based microfluidics (11), valves-based microfluidics (12), and droplet-based microfluidics (13).
Microfluidic Chips for Single-Cell Analysis
Microfluidic chip design helps in efficient sample isolation, cell sorting, lysis, and protein digestion, while liquid chromatography tandem mass spectrometry (LC–MS/MS) is the leading tool for protein identification and quantification (14). Single cell isolation can be achieved using droplet microfluidic chips wherein single cells are encapsulated within individual droplets, mainly by using an intersection of immiscible phases such as water-oil droplets in microchannels or through capillary tips (15). Further sorting and analysis of the cells is achieved by labeling of the single cells in droplets coupled with analytical methods such as biochemical reactions and fluorescence microscopy that enable real-time single cell detection. Recently, rapid droplet formation was achieved by constructing a micro cage array platform that contains multiple micropillars that can rapidly spread the oil phase through gaps between the pillars to trap droplets (16). Another method of achieving single cell isolation is by using microwells that can only accommodate one cell. The cell suspension is loaded onto the microwell array and single cells settle into each microwell due to gravity. Microwell arrays are a useful platform for studying the effects of drugs on single cells (17). Pang and associates investigated drug resistance of single-cell-derived tumor sphere based on an integrated microfluidic device. Single glioblastoma cells were isolated, captured, and then cultured into single cell derived spheres. Vincristine, the anti-cancer drug, was infused and co-cultured with the tumor-spheres. The study showed that tumor-spheres derived from smaller and more easily deformed tumor cells showed higher drug resistance (18).
While microwells employ physical boundaries to capture single cells, hydrodynamic trapping in microfluidic systems captures single cells by using specific fluid flows using cup-shaped micropillars (19) and S-shaped microchannels (20). Cup-shaped micropillar is one of the most used microstructures to isolate and trap single cells. Single-cell trapping is achieved by covering the gap in a micropillar that reduces the fraction of fluid streamlines that can flow into the trap. Microfluidic chips with cup-shaped micropillars have been shown to achieve an isolation efficiency greater than 95% for isolating single floating cancer cells (21).
Microfluidic Chip Coupled for Single-Cell Omics Analysis
Single-cell multi-omics, including transcriptomics, proteomics, metabolomics, and lipidomics, help to comprehensively understand cellular characteristics and cellular regulatory networks. The key requisite for the downstream analysis of single cells is the ability to be able to precisely manipulate single cells in microfluidic channels. Single-cell omics poses challenges to sampling techniques and MS sensitivity. Compared with single-cell metabolomics and lipidomics, single-cell proteomics has been more explored. Droplet microarray is one of the most suitable platforms for single-cell proteomics preprocessing, and has been used to understand cell phenotype differences and inter-relationships between functional networks (22). The droplets encapsulate the sample that provides compartments for extraction and digestion of proteins. This prevents overdilution of samples, and reduces the time for digestion (23). Comprehensive characterization of single cell proteome using sensitive MS is possible using single-cell sorting sample preparation from microfluidics, and provides detailed information of protein post-translational modifications as well as signal transduction-related functional proteins, such as transcription factors, kinases, receptors, and surface antigens.
Figure 1: Microfluidics coupled with MS for single cell proteomics, metabolomics, and lipidomics analysis. Figure adapted from reference (2).
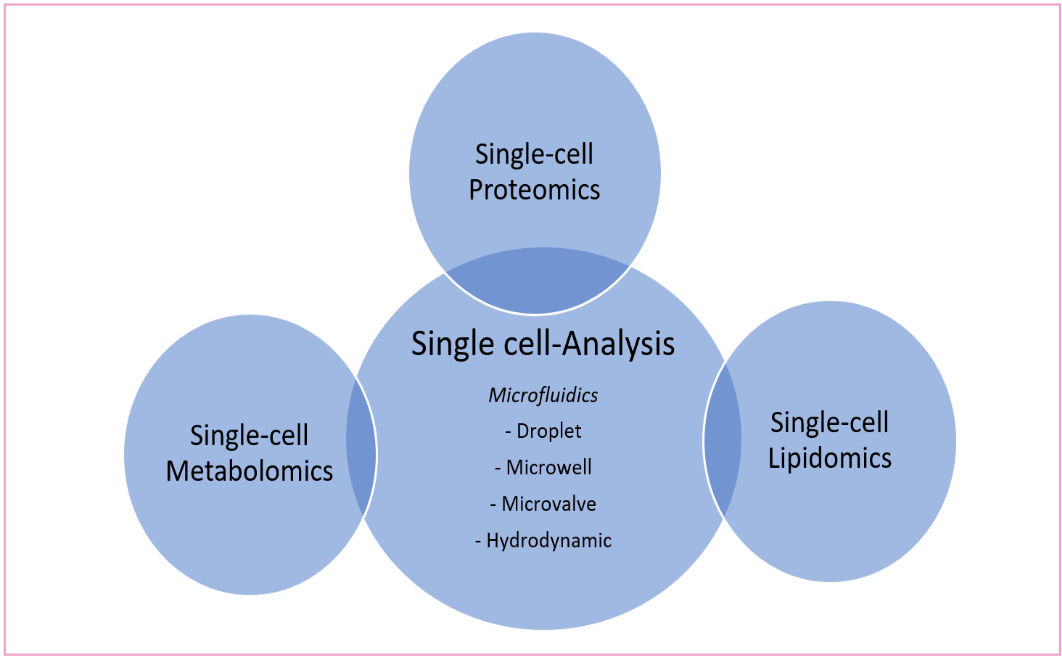
Microfluidic chips, composed of droplet microfluidics, arrays of microwells, arrays of micropillars, microchannels in special structures, and microvalves, can be used alone or in combination with bioanalytical assays to efficiently isolate and capture single target cells. Such a microfluidic platform has been used to sort immune cells, followed by lysis and digestion in centrifuge tubes for proteomic analysis. By using nano-LC–MS, 346 to 911 and 275 to 549 proteins were identified from 1000 and 100 THP-1 cells, respectively (24).
Gebreyesus and associates developed a streamlined and robust workflow that combined microfluidics and proteomic analysis based on data independent acquisition (DIA) MS for single-cell proteomic analysis, cell trapping, cell lysis, protein digestion, and peptide desalting to be performed on a single chip. The peptides collected from the chip were further analyzed using LC–MS/MS in DIA mode. 100 ± 131 protein groups were identified by the chip-DIA workflow from a single cell (25).
Due to the dynamic nature of metabolites to the environment, variations in metabolic status during analysis is a key area of concern. There are mainly two strategies for single cell metabolomic analysis; either analyzing intact cells directly by MS, or extracting metabolites from cells prior to MS detection (26,27). In recent work, a serpentine channel microfluidic chip was integrated with a pulsed electric field-induced electrospray ionization-high resolution MS (PEP-ESI-HRMS) to analyze single-cell metabolite (28). The asymmetric serpentine channel helped in continuously isolating single cells. Pulsed square wave electric filed was then applied to disrupt single cells and trigger ESI. Using the PEP-ESI-HRMS system, 80 cells could be analyzed in a minute, with about 120 metabolites identified per single cell. Microfluidic chips designed with narrow and long microchannels that allow only single cells to pass through can be used for cell delivery (29). Under a direct current voltage, single cells isolated in the microchannel can be lysed and ionized for MS analysis. The single-cell analysis platform can analyze about 40 cells per minute, and identify ≈100 metabolites from a single cell. The methodology led to the identification of 67 endogenous metabolites from single K562 cells (30). Similar with single cell metabolomic analysis, single-cell lipidome can also be analyzed using capillary ESI-MS to detect hundreds of lipid molecules with high resolution and accuracy. Single-cell metabolomics differs from lipidomics in the polarity of molecules; therefore a different solvent system is used for single-cell lipidomics. One of the challenges in single cell lipodomics is the complexity arising due to a large number of isomers of lipids. One possible solution is to integrate PB reaction with tandem MS to achieve the quantification of double bond position isomers in lipids (31,32).
The microfluidic MS-based single-cell omics has shown to have great potential in biomedical and clinical studies. It can help in revealing the heterogeneity of cells that is critical to study the progression of cancers, investigating the effect of drugs on the immune system as well as aid in understanding drug resistance during therapy (33,34). In addition, the characterization of single cell omics has the potential in the discovery of disease biomarkers.
Low Current Ionization in Studying Cell-to-Cell Heterogeneity
Ambient ionization refers to the ionization of unprocessed or minimally modified samples in their native environment, and it typically refers to the ionization of condensed phase samples in air. Ambient ionization MS techniques can be largely categorized into three classes based on their desorption method: liquid extraction, plasma desorption, and laser ablation. Ambient ionization MS techniques have been used for high-throughput analysis of clinical samples, including human biofluids, particularly for rapid therapeutic drug monitoring as well as to quantify drugs in cell cultures and tissue sections. This is mainly attributed to the ability to conduct the direct analysis of samples in their native environment without needing sample preparation (35).
Recent developments in ambient ionization mass spectrometry, such as sub-nanoampere ionization, have shown promise in studying cell-to-cell heterogeneity and its role in metabolomics. Sub-nanoampere ionization is a type of ambient ionization mass spectrometry that uses low currents to ionize samples. This approach has several advantages over traditional ionization methods, including the ability to analyze small numbers of cells along with the ability to analyze samples in their native environment. As an example, the pressure probe electrospray ionization MS with internal electrode capillary (IEC-PPESI-MS) can be integrated for high spatial resolution sampling and precise post sampling manipulation for single-cell analysis (36,37).
Another promising technology is paper spray ionization, which is a type of ambient ionization mass spectrometry that uses paper as a substrate for sample analysis. A paper triangle wetted with non-polar solvents such as hexane produces a spray of non-polar solvent droplets when a relatively low positive or negative voltage (0.8 ≈ 2 kV) is applied. The spray occurs at the tip of the paper, and is presumably due to field-assisted evaporation, while the solvent is transported as a result of capillary action through the micro-channels in the paper substrate (38).
This gentle method of ionization is applicable to a wide range of biological compounds including peptides, small nucleotides, phospholipids, and other compounds. This approach has several advantages over traditional ionization methods, including the ability to analyze small numbers of cells and the ability to analyze samples in their native environment by (i) low detection limits; (ii) low internal energy deposition; and (iii) compatibility with prior experiments that require aqueous or polar solvents, including prior chromatographic separations (39).
In a recent study, researchers used femto-flow electrospray ionization (ESI) with flow rates ranging from 240 fL min−1 to the low pico level (<10 pL min−1) using a submicron emitter tip and relay ESI configuration to analyze single cells. The researchers obtained high-quality mass spectra from single cells, which allowed them to identify metabolites not detectable using traditional methods (40).
Compared with nanoESI, femto flow ESI exhibits lower ion current intensities (> 2 orders of magnitude lower), ionization currents < 218 pA, and lower flow rates (> 3 orders of magnitude lower). The femto flow ionization technique allows he use of highly concentrated sample solutions in regular MS analysis, thereby removing the need for dilution when lower intensity ion beams are needed. Thus, for a non-buffered protein solution, the low flow ionization modes allow native charge states to be produced in an either charge-enhancing or charge-reducing manner. The measured flow rates and ionization currents then allow for the calculation of the size of initial charged nanodroplets. These techniques have the potential to revolutionize biopharmaceutical analysis by enabling the analysis of small numbers of cells in a cost-effective and efficient manner (41).
References
(1) Hodzic, E. Single-Cell Analysis: Advances and Future Perspectives. Bosn J. Basic Med. Sci. 2016, 16, 313. DOI: 10.17305/bjbms.2016.1371
(2) Zhang, D.; Qiao, L. Microfluidics Coupled Mass Spectrometry for Single Cell Multi-Omics. Small Methods 2023, 2301179. DOI: 10.1002/smtd.202301179
(3) Saviano, A.; Henderson, N. C; Baumert, T .F. Single-Cell Genomics and Spatial Transcriptomics: Discovery of Novel Cell States and Cellular Interactions in Liver Physiology and Disease Biology. J Hepatol. 2020, 73 (5), 1219–1230. DOI: 10.1016/j.jhep.2020.06.004
(4) Lim, B.; Lin, Y.; Navin, N. Advancing Cancer Research and Medicine with Single-Cell Genomics. Cancer Cell. 2020, 37 (4), 456–470. DOI: 10.1016/j.ccell.2020.03.008
(5) Zhang, L.; Vertes, A. Single-Cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity. Angew Chem. Int. Ed. Engl. 2018, 57 (17), 4466–4477. DOI: 10.1002/anie.201709719
(6) Katla, S. K.; Zhou, W.; Tavakoli, H.; Méndez, E. L. P.; Li, X. Portable in situ Temperature-Dependent Spectroscopy on a Low-Cost Microfluidic Platform Integrated with a Battery-Powered Thermofoil Heater. VIEW 2023, 4, 20220053.
(7) Dong, F.; Hao, S.; Zhang, S.; Zhu, C.; Cheng, H.; Yang, Z.; Hamey, F. K.; Wang, X.; Gao, A.; Wang, F.; Gao, Y.; Dong, J.; Wang, C.; Wang, J.; Lan, Y.; Liu, B.; Ema, H.; Tang, F.; Göttgens, B.; Zhu, P.; Cheng, T., Differentiation of Transplanted Haematopoietic Stem Cells Tracked by Single-Cell Transcriptomic Analysis. Nat. Cell Biol. 2020, 22, 630–639. DOI: 10.1038/s41556-020-0512-1
(8) Zhou, W. M.; Yan, Y. Y.; Guo, Q. R. et al. Microfluidics Applications for High-Throughput Single Cell Sequencing. J. Nanobiotechnol. 2021, 19, 312. DOI: 10.1186/s12951-021-01045-6
(9) Pihl, J.; Karlsson, M.; Chiu, D. T. Microfluidic Technologies in Drug Discovery. Drug Discov. Today 2005, 10 (20), 1377–1383. DOI: 10.1016/S1359-6446(05)03571-3
(10) Focaroli, S.; Mazzitelli, S.; Falconi, M.; Luca, G.; Nastruzzi, C. Preparation and Validation of Low-Cost Microfluidic Chips Using a Shrinking Approach. Lab Chip. 2014, 14 (20), 4007–4016. DOI: 10.1039/C4LC00679H
(10) Dai, X.; Shen, L. Advances and Trends in Omics Technology Development. Frontiers 2022. DOI: 10.3389/fmed.2022.911861
(11) Dendukuri, D.; Pregibon, D. C.; Collins, J.; Hatton, T. A.; Doyle, P. S. Continuous-Flow Lithography for High-Throughput Microparticle Synthesis. Nat. Mater. 2006, 5 (5), 365–369. DOI: 10.1038/nmat1617
(12) Berthier J; Brakke K. A;Berthier E. Open Microfluidics; Wiley, 2016.
Seemann, R.; Brinkmann, M.; Pfohl, T.; Herminghaus, S. Droplet Based Microfluidics. Rep. Prog. Phys. 2011, 75 (1), 016601. DOI: 10.1088/0034-4885/75/1/016601
(13) Zhu, F.; Liu, S.; Bai, X.; Liu, X.; Lin, B.; Lu, Y. Optimization Strategies of Covalent Organic Frameworks and Their Derivatives for Electrocatalytic Applications. VIEW 2023, 4, 20220033.
(14) Frey, C.; Pfeil, J.; Neckernuss, T.; Geiger, D.; Weishaupt, K.; Platzman, I.; Marti, O.; Spatz, J. P. Label-Free Monitoring and Manipulation of Microfluidic Water-in-Oil Droplets. VIEW 2020, 1, 20200101.
(15) Guo, X. L.; Wei, Y.; Lou, Q.; Zhu, Y.; Fang, Q. Manipulating Femtoliter to Picoliter Droplets by Pins for Single Cell Analysis and Quantitative Biological Assay. Anal. Chem. 2018, 90 (9), 5810–5817. DOI: 10.1021/acs.analchem.8b00343
(16) Lin, C. H.; Chang, H. C.; Hsu, C. H. A Microfluidic Platform for High-throughput Single-cell Isolation and Culture. J. Vis. Exp. 2016, 112, 54105. DOI: 10.3791/54105
(17) Pang, L.; Ding, J.; Ge, Y.; Fan, J.; Fan, S.K. Single-Cell-Derived Tumor-Sphere Formation and Drug-Resistance Assay Using an Integrated Microfluidics. Anal. Chem. 2019, 91 (13), 8318–8325. DOI: 10.1021/acs.analchem.9b01084
(18) Di Carlo, D.; Aghdam, N.; Lee, L. P. Single-Cell Enzyme Concentrations, Kinetics, and Inhibition Analysis Using High-Density Hydrodynamic Cell Isolation Arrays. Anal. Chem. 2006, 78 (14), 4925–4930. DOI: 10.1021/ac060541s
(19) Tan, W. H.; Takeuchi, S. A Trap-and-Release Integrated Microfluidic System for Dynamic Microarray Applications. Proc. Natl. Acad. Sci. USA 2007, 104 (4), 1146–1151. DOI: 10.1073/pnas.0606625104
(20) Tran, Q. D.; Kong, T. F.; Hu, D.; Marcos, L. R. H. Deterministic Sequential Isolation of Floating Cancer Cells Under Continuous Flow. Lab. Chip. 2016, 16 (15), 2813–2819. DOI: 10.1039/c6lc00615a
(21) Mashaghi, S.,; Abbaspourrad, A.; Weitz, D.A.; van Oijen, A.M. Droplet Microfluidics: A Tool for Biology, Chemistry and Nanotechnology. Trends Analyt. Chem. 2016, 82, 118–125. DOI: 10.1016/j.trac.2016.05.019
(22) Bellini, N.; Vishnubhatla, K.; Bragheri, F.; Ferrara, L.; Minzioni, P.; Ramponi, R.; Cristiani, I.; Osellame, R. Femtosecond Laser Fabricated Monolithic Chip for Optical Trapping and Stretching of Single Cells. Opt. Express 2010, 18, 4679–4688. DOI: 10.1364/OE.18.004679
(23) Bragheri, F.; Minzioni, P.; Martinez, V. R.; Bellini, N.; Paiè, P.; Mondello, C.; Ramponi, R.; Cristiani, I.; Osellame. R. Optofluidic Integrated Cell Sorter Fabricated by Femtosecond Lasers. Lab Chip. 2012, 12 (19), 3779–3784. DOI: 10.1039/c2lc40705a
(24) Gebreyesus, S. T.; Siyal, A. A.; Kitata, R. B. et al. Streamlined Single-Cell Proteomics by an Integrated Microfluidic Chip and Data-Independent Acquisition Mass Spectrometry. Nat. Commun. 2022, 13, 37. DOI: 10.1038/s41467-021-27778-4
(25) Spitzer, M. H.; Nolan, G. P. Mass Cytometry: Single Cells, Many Features. Cell 2016, 165 (4), 780–791. DOI: 10.1016/j.cell.2016.04.019
(26) Xu, S.; Xue, J.; Bai, Y.; Liu, H. High-Throughput Single-Cell Immunoassay in the Cellular Native Environment Using Online Desalting Dual-Spray Mass Spectrometry. Anal. Chem. 2020, 92 (24), 15854–15861. DOI: 10.1021/acs.analchem.0c03167
(27) Feng, J.; Zhang, X.; Huang, L.; Yao, H.; Yang, C.; Ma, X.; Zhang, S.; Zhang, X. Quantitation of Glucose-phosphate in Single Cells by Microwell-Based Nanoliter Droplet Microextraction and Mass Spectrometry. Anal Chem. 2019, 91 (9), 5613–5620. DOI: 10.1021/acs.analchem.8b05226
(28) Xu, S.; Liu, M.; Bai, Y.; Liu, H. Multi-Dimensional Organic Mass Cytometry: Simultaneous Analysis of Proteins and Metabolites on Single Cells. Angew Chem. Int. Ed. Engl. 2021, 60 (4), 1806–1812. DOI: 10.1002/anie.202009682
(29) Feng, J.; Zhang, X.; Huang, L.; Yao, H.; Yang, C.; Ma, X.; Zhang, S.; Zhang, X. Quantitation of Glucose-phosphate in Single Cells by Microwell-Based Nanoliter Droplet Microextraction and Mass Spectrometry. Anal. Chem. 2019, 91 (9), 5613–5620 DOI: 10.1021/acs.analchem.8b05226
(30) Capolupo, L. Single-Cell Lipidomics Reveals Organizing Principle of Cell Fate Decision. Nat. Rev. Mol. Cell Bio. 2023, 24, 377. DOI: 10.1038/s41580-023-00595-x
(31) Schober, Y.; Guenther, S.; Spengler, B.; Römpp, A. Single Cell Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Anal. Chem. 2012, 84 (15), 6293–6297. DOI: 10.1021/ac301337h
(32) Pattanayak, P.; Singh, S. K.; Gulati, M.; Vishwas, S.; Kapoor, B.; Chellappan, D. K.; Anand, K.; Gupta, G.; Jha, N. K.; Gupta, P. K.; Prasher, P.; Dua, K.; Dureja, H.; Kumar, D.; Kumar, V. Microfluidic Chips: Recent Advances, Critical Strategies in Design, Applications and Future Perspectives. Microfluid. Nanofluid. 2021, 25 (12), 99. DOI: 10.1007/s10404-021-02502-2
(33) Liu, D.; Sun, M.; Zhang, J.; Hu, R.; Fu, W.; Xuanyuan, T.; Liu, W. Single-Cell Droplet Microfluidics for Biomedical Applications. Analyst 2022, 147 (11), 2294–2316. DOI: 10.1039/D1AN02321G
(34) Takáts, Z.; Wiseman, J. M.; Gologan, B.; Cooks, R. G. Mass Spectrometry Sampling Under Ambient Conditions with Desorption Electrospray Ionization. Science 2004, 306 (5695), 471–473. DOI: 10.1126/science.1104404
(35) Gross, J. H. Direct Analysis in Real Time--A Critical Review on DART-MS. Anal. Bioanal. Chem. 2014, 406 (1), 63-80. DOI: 10.1007/s00216-013-7316-0
(36) Li, A.; Wang, H.; Ouyang, Z.; Cooks, R.G. Paper Spray for Direct Analysis of Complex Mixtures Using Mass Spectrometry. Chem. Commun. 2011, 47, 2811–2813. DOI: 10.1002/anie.200906314
(37) Li, A.; Wei, P.; Hsu, H. C.; Cooks, R. G. Direct Analysis of 4-methylimidazole in Foods Using Paper Spray Mass Spectrometry. Analyst 2013, 138, 4624–4630. DOI: 10.1039/C3AN00888F
(38) Liu, J.; Wang, H.; Manicke, N. E.; Lin, J. M.; Cooks, R. G.; Ouyang, Z. Development, Characterization, and Application of Paper Spray Ionization. Anal. Chem. 2010, 82 (6), 2463–2471. DOI: 10.1021/ac902854g
(39) Li, H.; Allen, N.; Li, M.; Li, A. Conducting and Characterizing Femto Flow Electrospray Ionization. Analyst 2022, 147 (6), 1071–1075. DOI: 10.1039/D1AN02190G
(40) Allen, N.; Li, H.; Cheung, A.; Xu, G.; Zi, Y.; Li, A. Femtoamp and Picoamp Modes of Electrospray and Paper Spray Ionization. Int. J. Mass Spectrom. 2021, 469, 116696. DOI: 10.1016/j.ijms.2021.116696
About the Authors
Anantdeep Kaur is a Postdoctoral Research Associate at the Biopharmaceutical Analysis Training Laboratory for the Department of Chemistry and Chemical Biology at Northeastern University, in Boston, Massachusetts.

Jahziel Chase is a third-year PhD candidate in the Department of Chemistry and Chemical Biology at Northeastern University, specializing in protein mass spectrometry. After graduating with a B.S. in Biochemistry from the University of Massachusetts Amherst in 2021, Jahziel Chase has dedicated his academic career to the field of mass spectrometry.

About the Editors
Jared R. Auclair is Interim Dean at the College of Professional Studies, Vice Provost Research Economic Development and Director of Bioinnovation at Northeastern University, in Boston, Massachusetts. He is also the Director of Biotechnology and Informatics, as well as the Director of the Biopharmaceutical Analysis Training Laboratory.

Anurag S. Rathore is a professor in the Department of Chemical Engineering at the Indian Institute of Technology in Delhi, India.


Accelerating Monoclonal Antibody Quality Control: The Role of LC–MS in Upstream Bioprocessing
This study highlights the promising potential of LC–MS as a powerful tool for mAb quality control within the context of upstream processing.
Common Challenges in Nitrosamine Analysis: An LCGC International Peer Exchange
April 15th 2025A recent roundtable discussion featuring Aloka Srinivasan of Raaha, Mayank Bhanti of the United States Pharmacopeia (USP), and Amber Burch of Purisys discussed the challenges surrounding nitrosamine analysis in pharmaceuticals.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)










