Accelerated Discovery and Quantitation of Lipids in Complex Extracts
Special Issues
The authors discuss analytical methods for lipidomics.
LC–MS-MS fractionation experiments or direct electrospray infusion can generate information-dependent MS-MS data triggered from a suite of triple-quadrupole selective scans or linear ion trap (LIT) scans. As shown in Figure 1, precursor ion (PIS), neutral loss (NL), multiple reaction monitoring (MRM), or enhanced MS LIT scanning can trigger an information-dependent enhanced product ion scan for MS-MS characterization of lipid structures.
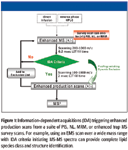
Figure 1
Faster quadrupole scanning and LIT scanning (up to 20,000 Da/s) combined with faster polarity switching and sensitivity improvements in enhanced product ion and MS3 scanning in LIT mode provide qualitative and quantitative strategies simultaneously. Figure 2 shows the coupling of scans for global lipid profiling on the AB SCIEX QTRAP 5500 system. Methods consisting of a suite of survey scans triggering multiple MS-MS scans yield in-depth lipid coverage in samples of wide dynamic range.
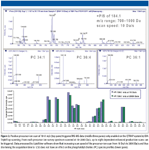
Figure 2
Targeted Multiple Precursor Ion Scanning by Direct Electrospray Infusion
A total of 60 combinations of PIS and NL multiplexed experiments can be acquired simultaneously for comprehensive quantitative analysis of lipid species by direct infusion. An example of data collected with negative mode multiple ion method consisting of multiplexed scanning of fatty acid acyl ions of varying saturation and carbon chain length, and diagnostic fragments for selected lipid classes, is shown in Figure 3. These data can be used directly to determine the abundance of lipids in a particular class from class-specific precursor ion experiments or to design subsequent MRM methods to quantify a subset of lipids of interest.
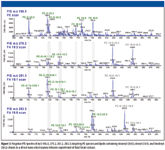
Figure 3
Measuring the Quantitative Response of Lipids to Class-Specific Internal Standards
Lipid class internal standards are essential to confirm, normalize, and quantitate corresponding lipid species that were detected with the MS approaches mentioned earlier (1). To study the linear dynamic range, a series of samples (three replicates for each concentration) consisting of the internal lipid class standards (held constant at 0.15 μM) and an increasing amount of total lipid extract from 10–3 to 102 μg were prepared from brain tissue. LIT enabled enhanced MS information-dependent acquisitions (IDA) triggered MS-MS methods were carried out and data were processed by LipidView software to evaluate the accuracy and precision of the quantitation. Figure 4 illustrates a linear response of representative lipids once corrected for their corresponding lipid-class specific internal standards across the calibration line over five orders of magnitude. Specifically for phosphatidyl choline (PC) and phosphatidyl ethanolamine (PE) lipids, the limits of detection are well in sub pmol level.
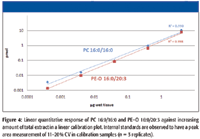
Figure 4
Post-Acquisition Data Processing for Lipid Identification and Quantitation
LipidView software is data processing software that identifies and quantifies lipid species from ESI-MS data. The software enables identification and characterization of lipid species through searching masses of parent and fragment ions in a database of characteristic fragments and neutral losses corresponding to parts of lipid molecules, and reports a numerical and graphical output for profiling various lipid molecular species, lipid classes, fatty acids, and long-chain bases (3). The software streamlines various steps, such as automated data processing method selection, lipid species identification, comprehensive isotope contribution removal, multiple internal standards-based quantification, visualization, and reporting of the results. A view of the user interface is shown in Figure 5 demonstrating the results workspace for reviewing the lipid profile data from a processed raw data file.
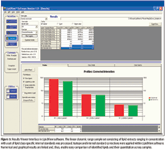
Figure 5
Improvements in Speed and Sensitivity for Lipidomics Applications
The AB Sciex QTRAP 5500 System's speed, selectivity, and sensitivity enable high multiplexing of QqQ scanning, and whole lipid extracts can be analyzed by either direct infusion of LC methods for in-depth lipid profiling — achieving both qualitative and quantitative data (with the use of synthetic lipid internal standards) in very short acquisition times. In addition to sensitivity gain, the MS system provides a polarity switching time of 50 ms, a linear accelerator trap scanning rate of 20,000 Da/s, and an LIT fill time of 50 μs. Lastly, Analyst 1.5 Software using the Scheduled MRM algorithm enables up to 2500 MRM transitions monitored in a single experiment for lipid quantitation in LC-based workflows.
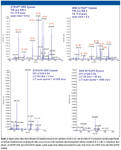
Figure 6
Conclusion
Scientists require versatility, sensitivity, specificity, and speed for high-throughput lipidome profiling of complex biological extracts by either direct infusion electrospray or LC–MS methods. For optimal results, data acquisition strategies should be coupled with automated data processing for lipid species identification and quantitation.
Brigitte Simons, Gary Impey, and Eva Duchoslav ar are with Applied Biosystems|MDS Analytical Technologies, Toronto, Ontario, Canada.
References
(1) M. Stahlman, C.S. Ejsing, K. Tarasov, J. Perman, J. Boren, and K. Ekroos, J. Chromatogr. B, Feb 2009, vol 37 epub.
(2) D. Schwudke, J. Oegema, L Burton, E. Entchev, J.T. Hannich, C.S. Ejsing, T. Kurzchalia, and A. Shevchenko, Anal. Chem. 78(2), 585–595 (2006).
(3) C.S. Ejsing, E. Duchoslav, J. Sampaio, K. Simons, R. Bonner, C. Thiele, K. Ekroos, and A. Shevchenko, Anal. Chem. 78(17), 6202–6214 (2006).

Polysorbate Quantification and Degradation Analysis via LC and Charged Aerosol Detection
April 9th 2025Scientists from ThermoFisher Scientific published a review article in the Journal of Chromatography A that provided an overview of HPLC analysis using charged aerosol detection can help with polysorbate quantification.
Analyzing Vitamin K1 Levels in Vegetables Eaten by Warfarin Patients Using HPLC UV–vis
April 9th 2025Research conducted by the Universitas Padjadjaran (Sumedang, Indonesia) focused on the measurement of vitamin K1 in various vegetables (specifically lettuce, cabbage, napa cabbage, and spinach) that were ingested by patients using warfarin. High performance liquid chromatography (HPLC) equipped with an ultraviolet detector set at 245 nm was used as the analytical technique.
Removing Double-Stranded RNA Impurities Using Chromatography
April 8th 2025Researchers from Agency for Science, Technology and Research in Singapore recently published a review article exploring how chromatography can be used to remove double-stranded RNA impurities during mRNA therapeutics production.












