What's the Issue with the LLOQ? — A Case Study
LCGC Europe
Two different methods of calculating the LLOQ disagree. Which, if either, is correct?
Two methods of calculating the lower limit of quantification (LLOQ) disagree. Which, if either, is correct?
Recently, a reader emailed me with a problem he was having determining the lower limit of quantification (LLOQ) for his method, which had a target LLOQ of 0.01 μg/mL for his analyte. He compared the LLOQ calculated using the International Committee on Harmonization guidelines (ICH) (1) with replicate injections of a reference standard and found that the two differed by more than an order of magnitude. He came to me to help him figure out what was wrong. The method was proprietary, and the reader needed to stay anonymous, so I've disguised things a bit, but this case study helps us to better understand how to evaluate a calibration curve.
The ICH (1) presents a formula to calculate what they call the quantitation limit (QL), but what most users call the limit of quantification (LOQ) or LLOQ:

where σ is the standard deviation of the response (the standard error [SE]) and S is the slope of the calibration curve. This is calculated easily from the regression statistics generated in Excel or your data system software. Let's see how this works.
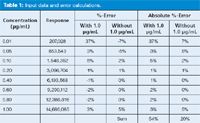
Table 1: Input data and error calculations.
Table 1 includes the initial data from the calibration curve. The user injected eight concentrations of his analyte, ranging from 0.01 to 1.0 μg/mL, generating the peak areas shown in the "Response" column of Table 1. I used Excel's regression tool to generate the regression statistics, part of which I've included in Table 2. These include the coefficient of variation (r2), the standard error of the curve (SE-curve), the y-intercept (intercept-coefficient), the standard error of the y-values (intercept-SE-y), and the slope of the curve (X variable). Calculated values for these variables are shown in the second two columns of Table 2, headed "With 1.0 μg/mL".

Table 2: Summary of Excel regression statistics.
The user used equation 1 with the standard error of the curve (SE-curve) and slope, and found that the LLOQ was predicted to be ~0.15 μg/mL (summarized as the first entry of Table 3). (Here I'll pause to remind you that I've rounded and truncated numbers in the tables for ease of viewing; if you try to repeat my calculations, your results may differ slightly.) Yet, when he injected n = 10 replicates of a 0.01 μg/mL solution, he found the percent relative standard deviation (%RSD) was 1.1% (last entry, Table 3), which he felt indicated the LLOQ was considerably lower than the 0.15-μg/mL prediction using the ICH technique. At this point he contacted me.
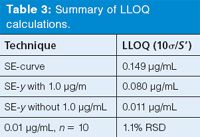
Table 3: Summary of LLOQ calculations.
Examine the Calibration Curve
The calibration curve shown in Figure 1(a) was supplied to me with the data set. You can see that the value of r2 = 0.9986 is excellent. The linear regression line is shown in blue; at first glance, this looks good too. However, a closer examination of the regression line shows that it is above the data points at low concentrations and below the data points at the high concentrations, passing through the data points at middle concentrations. This kind of behaviour tends to send up a caution flag for me because the higher concentrations tend to dominate the calculation. It is time to examine the data a little more carefully.

Figure 1: Plot of data of Table 1 with overlay of regression lines. Regression (a) including and (b) excluding the 1.0-μg/mL point.
Although it is part of the reporting requirements for most methods, we should be a little careful about putting too much confidence in values of r2. The reason for this is that the coefficient of variation is meant to be used with homoscedasic data; that is, data in which the standard deviation is approximately the same throughout the data range. Chromatographic data, however, are not homoscedastic, but heteroscedastic. The relative standard deviation (%RSD) tends to be constant throughout the range. In plain English, chromatographic data don't have, for example, standard deviations of ±1 ng/mL throughout the concentration range, but they might instead have ±0.1% RSD throughout the range. The coefficient of variation, r2, doesn't describe heteroscedastic data very well, so if we use r2 as our sole determinant of the goodness of a calibration curve, we may be misled. This all means that r2 = 0.9986 for these data does not guarantee that all is well.
Back to Table 1. I've used the regression equation to calculate the expected response at each concentration and compared this to the actual response to determine the percent error. These values are listed in the third column of Table 1 (%-error; with 1.0 μg/mL). You can see that the deviations from the expected values increase at lower concentrations, as expected, but they are also larger at high concentrations than in the middle of the curve. One technique to find out if there is a problem with the highest concentration is to drop it from the data set and repeat the calculations. I did this by dropping the 1.0-μg/mL point; the data are shown in column four of Table 1 (%-error, without 1.0 μg/mL). Notice how this reduces the deviations from the expected values. Also, the error increases at the lower concentrations, as expected, but is very small at higher concentrations (with the exception of 1.0 μg/mL). The regression results for the data without the 1.0-μg/mL point are shown in the last two columns of Table 2 (headed "without 1.0 μg/mL"). You can see that the SE-curve, SE-y, and y-intercept are all reduced by approximately an order of magnitude, yet r2 changes very little (0.9986 versus 1.000). In Figure 1(b), I've plotted the revised regression line, which visibly fits the data better than the original if the 1.0-μg/mL point is ignored. Another way to evaluate these differences is to compare the absolute values of the %-error, as shown in the last two columns of Table 1. The sum of these absolute values is shown at the bottom. Notice that eliminating the 1.0-μg/mL point from the regression calculation reduced the total by more than 2.5-fold from 54% to 20%. This is definitely a better fit of the data.
An additional way to visualize the data is shown in Figure 2, where I've taken just the lower (Figure 2[a]) and higher (Figure 2[b]) portions of the concentration curve and expanded the scale. Now the original regression (blue line) is obviously an inferior fit to the revised one (red line) at both ends of the scale.

Figure 2: Expanded sections of Figure 1(a) (blue) and 1(b) (red): (a) 0.01â0.2 μg/mL region, (b) 0.6â1.0 μg/mL region.
At this point, it might be interesting to determine what the problem is with the 1.0-μg/mL point, but I don't have any additional information to help me with this task. It would be nice to make several replicate injections to be sure the 1.0-μg/mL data point isn't an outlier. If the problem persists over replicate injections, a new preparation of the standard should be checked to eliminate the possibility of formulation errors. Another possibility is that the peak is large enough to cause a slightly nonlinear behaviour of the detector, which often happens as the detector signal nears its upper limit. In any event, I think it is prudent to exclude this point from the regression without further indications that it should be included as a valid point.
Another question that often comes up is whether the calibration curve should be forced through x = 0, y = 0 or not. This is a simple test that was discussed in an earlier "LC Troubleshooting" column (2). If the value of the y-intercept calculated from the regression process is less than the standard error of the y-intercept, it means that the y-value is within 1 standard deviation (SD) of the 0,0 point. Most statistical tests will tell you that there is no statistical difference between a point <1 SD from the mean and the mean, so the curve can be forced through zero. How do you check this? The data are in the Excel regression summarized in Table 2 on the line labelled "intercept." The "coefficient" column lists the calculated value of the y-intercept, so if this is less than the standard error (SE-y), you can force the curve through zero. You can see that in both cases (with or without 1.0 μg/mL included), the y-intercept is greater than the standard error, so the curve should not be forced through zero.
Double-Check the Calculations
Now that we've decided to exclude 1.0 μg/mL from the regression calculations, let's see why the ICH method predicted such a large LLOQ. When I tried to reproduce the user's results, I found the problem. He was using the standard error of the curve (SE-curve, line 2 of Table 2) instead of the standard error of the y-intercept. The SE-curve value represents the variability around the regression curve throughout the whole range of the curve. But for determination of the LLOQ, we want to use the standard error in that region instead, so SE-y is more appropriate. Otherwise we often find that the variability of the larger concentrations overpowers the variability of the lower ones and gives an unrealistically high value of the LLOQ. When I used the SE-y value with equation 1, the LLOQ was reduced by approximately twofold with the original dataset (0.15 versus 0.08 μg/mL), as shown in the first two lines of Table 3. When the SE-y of the revised calibration curve (without 1.0 μg/mL) is used, the predicted LLOQ drops to 0.011 μg/mL. As mentioned above, the revised calibration curve generates values of SE-curve and SE-y that are approximately an order of magnitude smaller than the original data set (Table 2).
The predicted LLOQ that we just calculated using the ICH method is not sufficient, however. The ICH document (1) clearly states, "the limit should be subsequently validated by the analysis of a suitable number of samples known to be near or prepared at the quantitation limit." In the last line of Table 3, you can see that the n = 10 replicate injections at 0.01 μg/mL gave imprecision of 1.1% RSD, an excellent value at the LLOQ for most methods. This strongly suggests that the method will perform adequately at the desired LLOQ of 0.01 μg/mL of the target analyte.
Summary
This data set has served as a good example of how easy it is to misinterpret the results of a calibration curve. We saw that the value of r2 can be misleading about how good the calibration curve is. It was shown that it is useful to examine both a visual and tabular expression of the data. The original plot (Figure 1[a]) suggested that the highest concentration might be biasing the regression, and when this point was eliminated, the new trend line (Figure 1[b]) fits all the other points better. Expanding the scale on the plots (Figure 2) also helped to get a better picture of what is happening. Comparing the sum of absolute values of the deviations of experimental data points from those calculated from the regression is a simple way to see if a new data treatment reduces the overall error. In the present case, error was reduced by more than 2.5-fold simply by dropping the highest concentration point (Table 1).
When using estimating techniques, such as the ICH method used here, it is imperative to use the correct coefficients or the wrong conclusions may be drawn. Fortunately, the user noticed that something was wrong and searched for further help. If, instead, he believed the calculations, he might have discarded a good method or spent unnecessary time trying to improve an already acceptable method. Finally, regression curves, percent-error tables, and data plotting techniques are merely tools to help us understand the data better. When it comes to determining the LLOQ, there is nothing that can compare with the measured performance from multiple injections at the target LLOQ.
John W. Dolan is the vice president of LC Resources, Walnut Creek, California, USA. He is also a member of the LCGC Europe editorial advisory board. Direct correspondence about this column should go to "LC Troubleshooting", LCGC Europe, 4A Bridgegate Pavilion, Chester Business Park, Wrexham Road, Chester, CH4 9QH, UK, or e-mail the editor-in-chief, Alasdair Matheson, at amatheson@advanstar.com
References
(1) Validation of Analytical Procedures: Text and Methodology Q2(R1), International Conference on Harmonization, Nov. 2005, http://www.ich.org/LOB/media/MEDIA417.pdf.
(2) J.W. Dolan, LCGC North Am. 27(3), 224–230 (2009).

New Study Reviews Chromatography Methods for Flavonoid Analysis
April 21st 2025Flavonoids are widely used metabolites that carry out various functions in different industries, such as food and cosmetics. Detecting, separating, and quantifying them in fruit species can be a complicated process.
Quantifying Terpenes in Hydrodistilled Cannabis sativa Essential Oil with GC-MS
April 21st 2025A recent study conducted at the University of Georgia, (Athens, Georgia) presented a validated method for quantifying 18 terpenes in Cannabis sativa essential oil, extracted via hydrodistillation. The method, utilizing gas chromatography–mass spectrometry (GC–MS) with selected ion monitoring (SIM), includes using internal standards (n-tridecane and octadecane) for accurate analysis, with key validation parameters—such as specificity, accuracy, precision, and detection limits—thoroughly assessed. LCGC International spoke to Noelle Joy of the University of Georgia, corresponding author of this paper discussing the method, about its creation and benefits it offers the analytical community.










