Visualizing the Chemical Composition of Complex Samples
LCGC North America
How does one decipher and tabulate thousands of chemical formulas and relative abundances without losing the important details?
The materials examined by mass spectrometrists may have hundreds to thousands of unique components in a wide concentration range. The spectra are very complex; to even a trained eye, two mass spectra of related but different samples are virtually indistinguishable. So how does one decipher and tabulate thousands of chemical formulas and relative abundances without losing the important details?
As mass spectrometrists, we regularly encounter complex samples. Frequently, we're asked to isolate, identify, and quantify analytes from complex matrices like body fluids or tissue, soil, water, agricultural commodities, or process streams. When performing examinations in the presence of impurities and chromatographic or spectral interferents, we apply a variety of separation tools and protocols. Eventually, we produce an aliquot of material that is, we expect, clean enough to provide unambiguous results. Yet even then, the extreme lengths we took to produce that aliquot might have failed to deliver us from our dilemma. At such times, we resort to our most sensitive means of analysis — highly selective mass spectrometry (MS) techniques like ion mobility or multiple reaction monitoring (MRM) — to eliminate isobaric interferences.
For another group of researchers, though, the whole sample is the analyte. For them, extracting and analyzing a part of the sample is of no use, and only a compositional characterization of the entire sample provides the information required to address the experiment's goals. The materials examined by these scientists may have hundreds to thousands of unique components, in a wide concentration range. The spectra are very complex. Even to a trained eye, two mass spectra of related but different samples are virtually indistinguishable, and it is the identification and relative quantification of their components that tells the story. Nevertheless, looking at tabulations of thousands of chemical formulas and relative abundances can quickly glaze the eyes of any observer, while important details get lost in the blur of numbers.
The MS analysis of petroleum is a case in point. The March 2008 installment of this column (1) described in detail the techniques developed by Alan Marshall, Ryan Rodgers, and their research team from the National High Magnetic Field Laboratory (NHMFL) at Florida State University (Tallahassee, Florida). The petroleum samples this group handles are likely some of the most complex materials ever analyzed, often yielding mass spectra that include nearly 30,000 peaks. Using high performance instruments and specialized software tools to analyze these perplexing spectra, they can assign a chemical formula and relative abundance to each of those peaks.
Yet even the ability to perform such a remarkable feat is not answer enough. The development of visualization tools that condense tables of chemical formulas into easily interpretable plots and images useful to pipeline operators and refinery process engineers plays a major role in the analysts' success. With such pictures, they can readily convey the composition of a single sample and compare samples from different sources or processing conditions.
Over the course of more than 20 years of research in petroleum analysis, the NHMFL scientists have become widely recognized as world leaders in this field, with an extensive publication record covering their analysis and visualization techniques. Now, the introduction of a new generation of affordable, high-resolution, high-accuracy mass spectrometers makes possible the routine generation of high quality mass spectral data outside the confines of a national laboratory. Thus, scientists working in domains other than petroleum have investigated ways to apply these petroleum-analysis techniques to their samples, some of which can be as complex as petroleum. In this column, we will meet three such scientists, from industrial, academic, and government research groups, who work with such diverse samples as formulated products, swamp water, and atmospheric aerosols.
When a Defective Analysis Really Isn't
A key concept in the analysis and characterization of complex samples is the mass defect. Based in nuclear physics, the mass defect is defined as the difference between the mass of an atom and the sum of the masses of the individual subatomic particles that comprise it. Because of the binding energy that holds the atom together, its mass is always less than the total unbound masses of the particles. In molecular terms, the mass defect is the difference between the sum of the monoisotopic mass of the individual atoms and their mass number, which is the sum of the number of nucleons in the nuclei of those atoms. As described in the earlier column (1), the "exact" monoisotopic mass of the water molecule, consisting of two atoms of hydrogen (1H) and one of oxygen (16O) is 2 × 1.0078 + 15.9994 = 18.0106 u. The sum of nucleons is 18, so the mass defect of the H2O molecule is 0.0106 u. The IUPAC standard assigns the unified atomic mass unit to be 1/12 the mass of a 12C atom; thus, 12C has an atomic mass of 12.0000 u and a mass defect of zero.
Given that every isotope of every atom has a slightly different mass defect, each chemical composition also has a unique defect. If two compounds are related by some chemical difference, then their mass defects differ by the mass defect of the difference formula. For example, methane (CH4; 16.0312 u) and ethane (C2H6; 30.0468 u) differ by a methylene (CH2; 14.0156 u). Their mass defects differ by the mass defect of methylene, 0.0156 u. In recent years, this property has been applied in drug metabolite analysis to perform mass defect filtering of spectra. This technique, developed by Zhang, Zhu, and colleagues at Bristol Myers Squibb (2,3), simplifies liquid chromatography (LC)–MS spectra by removing all peaks that do not fall within a narrow mass defect window relative to the substrate. The defect window used is characteristic of a particular metabolic transformation, such as oxidation. The peaks remaining after filtering are likely to be caused by such oxidative metabolites.
The same property applies for any series of homologous compounds: The mass defect difference between any two successive members of the series is the mass defect of the repeating unit of the series. In 1963, Kendrick (4) recognized that this property could prove useful when analyzing petroleum by MS. By scaling the mass axis of a hydrocarbon spectrum by a factor equal to the ratio of the nominal mass of CH2 to its exact mass (14/14.01565 ≈ 0.9989), the mass of CH2 in the resultant spectrum is exactly 14.000, and all peaks of a given homologous series are exactly 14 u apart.
As an additional consequence of this rescaling, the mass defects of successive members of the same homologous series are precisely the same because, on this scale, the mass defect of the repeating methylene unit is zero. Two hydrocarbon series that differ by one degree of unsaturation (that is, a chemical difference of H2) are offset by the scaled mass defect of H2. Likewise, when the difference results from the presence of one or more non-CH atoms in the core formula of the hydrocarbon, all pairwise members of the two homologous series (those of the same CH2 count) differ by the same mass defect. Rodgers and colleagues exploit this effect. They create Kendrick dot plots, or images of Kendrick mass defect vs. Kendrick mass. Thus, in a two-dimensional dot plot, the peaks of each homologous series form a horizontal row of dots, each row offset from the others by a difference in unsaturation or core composition.
Such dot plots serve as the basis of the algorithm for assigning chemical composition to these complex spectra. By assigning a de novo chemical formula to one dot (peak) in a row, all other dots in the same row can also be assigned a chemical formula simply by adding multiples of CH2. After one row is assigned, the algorithm jumps to another row using the Kendrick H2 mass defect, because all the dots in the latter row are related to all the dots in the first row by a chemical difference of H2. Thus, the assignment procedure works its way through the entire array of dots, one row at a time, first by assigning a de novo composition to one dot, then tracking through all related dots by chemical formula differences.
The de novo assignment of an initial composition depends on high mass accuracy. The inherent mass accuracy of the instrument must generally be improved upon by internal recalibration of the spectrum using a series of target peaks of known composition spaced throughout the spectrum. The NHMFL group has developed several recalibration methods, the most recent of which allows a low parts-per-billion mass accuracy throughout the spectrum (5). At this level of accuracy, a peak of relatively low molecular weight can be assigned a composition unambiguously.
After a chemical composition is assigned, a petroleum sample can be grouped into subsets by heteroatom content (chemical class) and by degree of unsaturation (the double bond equivalent, or DBE). The DBE is easily computed from the chemical formula
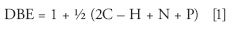
where the letters represent the counts of the respective atoms, and the lowest valence of N and P is assumed. The DBE is a measure of the number of rings plus double bonds, and for petroleum samples, higher DBE values are generally associated with highly condensed and aromatic ring systems. In addition to DBE, the chemical formula provides other measures: atom counts and ratios of atom counts. In hydrocarbons, the carbon count is roughly equivalent to molecular weight, so it is common to create smoothed images of DBE vs. carbon number for members of the same hydrocarbon class. By color coding such images, using a gradient based on relative abundance, the image readily imparts a visual indication of the ranges of both DBE and molecular weight, as well as the mean and standard deviation of each. Comparing two images (such as before and after catalytic cracking to reduce molecular weight and increase the concentration of low-boiling aromatics) should show a shift of the image along both axes to a lower carbon number and a lower DBE.
Ratios of atomic compositions are used to produce so-called van Krevelen plots. A typical 2D van Krevelen plot uses the H:C ratio as the ordinate and a heteroatom:C ratio (O:C, N:C) as the abscissa. On these plots, compounds of related composition form rows of dots along the horizontal, vertical, or diagonals. For example, in a H:C vs. O:C van Krevelen plot, compounds related by hydrogenation or dehydrogenation form vertical lines; compounds differing by oxygen count (oxidation or reduction) form horizontal lines. Compounds related by decarboxylation, hydration or dehydration, or methylation or demethylation, form various diagonal lines.
Solving Formulation Problems Using Graphics
Robert Strife of Procter & Gamble (P&G, Cincinnati, Ohio) is responsible for the characterization of "known unknowns," which are formulated materials whose constituents are all known commercial compounds, but whose identities and relative amounts are unknown to the investigator. In an oral presentation at the September 2010 CoSMoS conference on small molecule science (6), Strife presented two examples of how "mass mapping" (his term for two-dimensional plotting of chemical compositions) allows him to diagnose and solve problems with complex raw materials destined for use in P&G's food or cosmetics products.
In one application, polyglycerol esters (PGEs), which consist of a polyglycerol backbone esterified with coconut-oil fatty-acid esters (compositions C12, C14, C16, C18, C18:1), are used as a "green" raw material to form emulsions. To perform adequately in this application, the number of glycerol units and the degree of esterification must fall within an acceptable range and be present in appropriate concentrations. The number of combinations of glycerol units, esterification positions, fatty acid types, and multiple adducts makes for spectra of astounding complexity: For a polyglycerol with 5 glycerol units in the backbone (a "G5"), between 1 and 7 esterification sites, and the 5 fatty acids listed above, there are 774 combinations. Add to this the possibility of G1–G4, or higher, oligomers, a potential contaminant resulting from cyclization of a glycerol subunit and loss of H2O, and molecular ions from electrospray ionization with H+, Na+, and NH4+ adducts, and the spectra become forests of peaks (Figure 1). In such spectra, it is impossible to discern by eye anything more than gross differences between good or bad samples. Often, difference between a material that performs successfully and one that does not is subtle variation in the relative amounts of each of the polyglycerol esters.
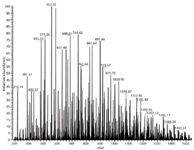
Figure 1: Infusion ESI+ Fourier transform orbital trapping MS spectrum of a polyglycerol polyester.
To make sense of all this, Strife adapted the Kendrick mass defect plot. Using C3H6O2 as the Kendrick "basis" (rather than CH2) results in a normalization of the mass axis to one in which molecules with successive glycerol backbone units are exactly 74 u apart. Along the y axis, the mass defect scale is also normalized to glycerol. Thus, every polyglycerol of the same ester composition, differing only by glycerol count, manifests the same mass defect and falls on a horizontal line. Cyclic glycerol impurities fall on horizontal lines as well, offset from the noncyclic PGEs by the scaled mass and mass defect of water. For a given glycerol count, differences in ester chain length fall on diagonal lines. Finally, the mono-, di-, triesters and higher form neatly separated groups (Figure 2). By transforming and mapping these complex spectra in this way, Strife converts an indescribably complex spectrum into easily interpretable maps by which one can readily discern differences in composition or the presence of unacceptable levels of impurities.

Figure 2: A Kendrick mass defect map of polyglycerol polyester, normalized to C3H6O2.
In a second example, Strife borrows a visualization method from the proteomics field. In a 2009 paper (7), Roman Zubarev and colleagues proposed a two-dimensional plot with the ordinate representing a normalized isotopic shift (NIS) and the abscissa a normalized mass defect (NMD). These two quantities can be calculated from careful measurements of isotope clusters in experimental spectra or from calculations based on chemical formulas, as follows
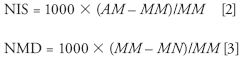
where AM is the average (or chemical) mass, MM is the monoisotopic mass, and MN is the nominal molecular mass, respectively. The NIS is effectively the normalized difference between the monoisotopic mass and the centroid of the molecular ion cluster's isotopic envelope. Thus, it generally increases with increasing molecular weight (as the abundance of 13C-containing isotopic peaks increase relative to the monoisotopic 12C peak) and, more importantly, increases dramatically with the addition of atoms with significantly abundant higher isotopes, such as S, Cl, or Br. The NMD axis varies with degree of unsaturation as well as heteroatom content. In his paper, Zubarev used this 2D plot to illustrate how subtle differences in the composition of a peptide could result in large shifts along one or both axes of the plot.
Strife illustrated how to adapt this "Zubarev plot" to map compositional differences in complex mixtures. As an example, he created a plot of an artificial mixture of polyglycerol esters, alkyl ethoxylates (EO), and alkyl ethoxylates phosphate mono- and diesters. As shown in Figure 3, the various classes are clearly separated, with related materials forming linear clusters of dots. The presence of an impurity would be glaringly obvious as an out-of-place dot on such a map.

Figure 3: A "Zubarev plot" demonstrating clustering and class separation of polyglycerol polyesters (PGE) and various alkyl polyethoxylate (EO) and EO phosphate ester species.
Mapping the Complexity of Natural Organic Material
Rachel Sleighter, research manager for the group led by Patrick Hatcher at Old Dominion University in Norfolk, Virginia, manages a large research team studying the composition of natural organic matter in dissolved water, soil and sediment, aerosol, fossilized material, and so forth. The most difficult aspect of this work, Sleighter says, is relating the organic characterization to other environmental observations in a meaningful way (for example, water quality and pollution transport, air mass trajectories, or carbon cycling between sources and sinks). The group's primary tool for molecular-level characterization is electrospray ionization, Fourier-transform ion cyclotron resonance mass spectrometry (ESI-FT-ICR-MS). Other tools, such as gas chromatography–mass spectrometry (GC–MS), high performance liquid chromatography (HPLC), nuclear magnetic resonance (NMR) spectroscopy, and Fourier-transform infrared (FT-IR) spectroscopy are used to characterize specific components or for bulk-level characterization.
The FT-ICR-MS spectra include thousands of individual peaks that are carefully calibrated to ensure high mass accuracy. Using a molecular formula calculator developed by the NHMFL team and guided by knowledge of the expected composition of the natural organic matter, Sleighter's group can assign a chemical formula to the majority of the peaks in the spectrum. Given the formulas, Sleighter can apply the visualization methods developed for petroleum samples. Compared to petroleum, natural organic matter is generally highly oxygenated, so Van Krevelen plots of H:C vs. O:C composition tend to be the most frequently used. On such a plot, various chemical classes within natural organic matter, such as humic or fulvic acids, cellulosic materials, and lignins form distinct clusters. Comparison of plots of samples that have undergone chemical or biological transformation reveal characteristic shifts associated with the degree of degradation.
To better understand these changes, the Hatcher group has recently begun using multivariate statistics to compare large numbers of samples (8). "Because our FT-ICR-MS analyses translate into datasets containing thousands of molecular formulas, it has been a challenge to effectively compare a large number of samples and to determine subtle differences," Sleighter says.
Multivariate statistical methods like principal component analysis (PCA) identify groups of formulas that seem to indicate important differences between samples. Sleighter commented on those methods when used in combination with Van Krevelen plots that map the formula differences into chemical class differences. "We can design new experiments in order to understand these components better or to correlate them with other measurements and environmental observations," she says (Figure 4).

Figure 4: Principal component analysis on a subset of peaks detected in at least one of 38 DOM samples from various terrestrial, estuarine, and marine locations and analyzed by ESI-FT-ICR-MS: (a) the biplot of the formula loadings (variables) in the PCA indicate that high positive PC1 and PC2 values correlate to estuarine and marine samples while high negative PC1 and PC2 values correlate to terrestrial samples; (b) the corresponding van Krevelen diagram of the assigned formulas, colored according to location on the PCA biplot. (Adapted from reference 8.)
Mass Defect Folding to Reduce Complexity
In a final example of complexity reduction, the work of Julia Laskin, chief scientist in the Chemical and Materials Sciences Division of the Pacific Northwest National Laboratory (Richland, Washington), illustrates how the application of multiple orders of mass defect "folding" can reduce an extremely complex spectrum to a small set of points. One area of Laskin's research relates to the chemical characterization of the large organic polymers that constitute a significant fraction of secondary organic aerosols (SOA) and the study of their effects on physical properties of aerosols relevant to climate change. In a recent paper (9), Laskin and colleagues describe how successive applications of mass defect scaling transformation can reduce a complex spectrum to a set of a few dots on a two-dimensional mass defect plot.
In an ordinary mass defect plot, a single "basis formula" is used to rescale the mass and mass defect axes. As we have seen, such rescaling results in a plot where each homologous series forms a horizontal row of dots, incremented by the nominal mass of the basis formula. Series that differ by degree of unsaturation or core composition form additional rows spaced along the mass defect axis. In this first-order plot, each dot represents a single monoisotopic peak in the mass spectrum. In Laskin's work, application of a second-order mass defect scaling reduces each row of dots to a single dot. In this second-order plot, the abscissa is replaced by the original mass defect axis (the ordinate in the first order plot), and a new scaling of the ordinate is performed by scaling the first-order defect by a new defect for a second-basis formula. By factoring out the first-order defects in this way, each homologous series collapses into a single dot. This can be extended to a third order using yet another repeat formula as basis, and so on. Each of these folding steps reduces the complexity dramatically, at the loss of the detailed information factored out by the folding.
Using a petroleum spectrum with nearly 13,000 monoisotopic peaks as an example, the first-order mass defect plot (using CH2 as a basis) is a dark blob of unresolved dots. The second-order of folding uses both CH2 and H2 as bases, reducing the 13,000 dots into 25 horizontal lines containing a total of 480 dots. By folding both the chain length (CH2) and degree of unsaturation (H2) into the ordinate, each of the 25 lines now represents a specific chemical class (containing a core composition of N1, N2, NO, O, O2, S, and so forth). Every dot along a given horizontal class line represents a homologous series of the same degree of unsaturation. Folding a third time, using oxygen (O) as a basis, reduces the 480 dots into only 25 dots. Now each horizontal line represents a class of a specific, non-oxygen, heteroatom composition, with successive horizontal dots shifted by the effect of zero, one, two, three, or more oxygen atoms. From a spectrum of 13,000 points, these successive foldings result in a plot of only 25 points, yet the complete heteroatomic composition of the sample is immediately comprehensible. (Figure 5) For the analysis of petroleum, natural organic matter, and other such materials whose composition is highly complex but also highly regular, this reduction of complexity should be of great utility in both the assignment of chemical composition and comparison between samples.
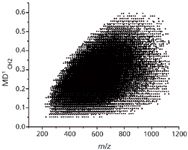
Figure 5: (a) A Kendrick mass defect plot of a crude oil sample. The mass defect on a CH2 basis is plotted versus the Kendrick m/z, also based on CH2. Each of the nearly 13,000 dots represents a single monoisotopic peak in the mass spectrum. (b) The same spectrum, after three successive mass defect folding steps. The abscissa is the third order mass defect, based on CH2, H2, and O, plotted vs. the second order defect based on CH2 and H2. In this plot, each horizontal row represents a heteroatom composition class as labeled. Successive dots in each row represent 0, 1, 2 . . . additions of an oxygen atom to the core composition. (Adapted from reference 9.)
David Stranz is the president of Sierra Analytics, Inc., in Modesto, California. He obtained a Ph.D. in physical chemistry from the University of Maryland, College Park (College Park, Maryland). He worked as an analytical chemist for Shell Chemical Company and at the E. I. du Pont de Nemours and Co. Experimental Station, and developed mass spectrometry software at Hewlett-Packard, Fisons Instruments, and Micromass before cofounding Sierra Analytics in January 1997. He can be contacted at David_Stranz@MassSpec.com.

David Stranz
Michael P. Balogh "MS — The Practical Art" Editor Michael P. Balogh is principal scientist, MS technology development, at Waters Corp. (Milford, Massachusetts); a former adjunct professor and visiting scientist at Roger Williams University (Bristol, Rhode Island); cofounder and current president of the Society for Small Molecule Science (CoSMoS) and a member of LCGC's editorial advisory board.

Michael P. Balogh
References
(1) M.P. Balogh, LCGC 26(3), 262–276 (2008).
(2) M. Zhu, L. Ma, H. Zhang, and W.G. Humphreys, Anal. Chem. 79(21), 8333–8341 (2007).
(3) H. Zhang, D. Zhang, K. Ray, and M. Zhu, J. Mass Spectrom. 44, 999–1016 (2009).
(4) E. Kendrick, Anal. Chem. 35(13), 2146–2154 (1963).
(5) J.J. Savory et al., Anal. Chem. 83(5), 1732–1736 (2011).
(6) R.J. Strife, "Normalized Mass Mapping to Characterize 'Green' Raw Materials Analyzed by Fourier Transform Orbital Trapping Mass Spectrometry", paper presented at CoSMoS 2010, Portland, Oregon, 2010.
(7) K.A. Artemenko et al., Anal. Chem. 81(10), 3738–3745 (2009).
(8) R.L. Sleighter, Z. Liu, J. Xue, and P.G. Hatcher, Environ. Sci. Technol. 44(19), 7576–7582 (2010).
(9) P.J. Roach, J. Laskin, and A. Laskin, Anal. Chem. 83(12), 4924–4929 (2011).

New Method Explored for the Detection of CECs in Crops Irrigated with Contaminated Water
April 30th 2025This new study presents a validated QuEChERS–LC-MS/MS method for detecting eight persistent, mobile, and toxic substances in escarole, tomatoes, and tomato leaves irrigated with contaminated water.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)









