Variable Selection and Reduction in Multivariate Calibration and Modelling
LCGC Europe
In multivariate calibration and modelling a model is built between a large data matrix containing many variables for each sample and the property of the sample.
In multivariate calibration and modelling a model is built between a large data matrix containing many variables for each sample, such as a spectrum or a chromatogram, and a property of the samples, such as a given activity. Variable selection or reduction in the large matrix is required, or can be recommended, to obtain better and simpler models.
In pharmaceutical data analysis multivariate calibration and modelling are often used to build predictive models. Many different modelling techniques can be used. However, these techniques usually require either a variable selection step to limit the number of variables to build the model or create so called latent variables to build simple models.
Many variable selection methods have been applied in the literature. An overview of some of the most frequently applied will be discussed in this article In cases where latent variables are used, the models with latent variables based on the entire data set are usually not as good as when uninformative variables are eliminated first. Different possibilities exist to reduce the number of variables used in the calculation of the latent variables. The goal of feature selection or reduction is to select the most important but also to build simpler methods with a better predictive performance than when applying the entire data set.
In several data handling approaches of pharmaceutical data sets either multivariate calibration or multivariate modelling is applied. In multivariate calibration a multivariate model is built between, for example, a near infrared (NIR) spectrum, a Raman spectrum, or an entire chromatogram (e.g., a fingerprint) of the samples, on the one hand, and a property or an activity on the other. Activities and properties that are considered are, for example, the water content (from NIR spectra), active compound concentration (from spectra), antioxidant activity, cytotoxic activity, or anticancer activity (from spectra or chromatograms). The general model can be represented as
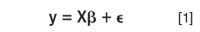
where y is the vector of the observations (concentrations, activities...), X is the matrix of the independent variables (absorbance at given wavelengths in a spectrum or detector signal at a at given time in a chromatogram), β the vector of the regression coefficients to be estimated and ζ t the error vector (1). The model is then used to predict the modelled property for new samples from its measured spectrum or chromatogram.
In multivariate modelling an activity or a retention time is modelled as a function of molecular descriptors, which represent chemical structure properties. These models are called quantitative structure-activity relationship (QSAR) and quantitative structure-retention relationship (QSRR) models, respectively. Activities can be those mentioned above, but also protein-binding capacities or enzyme-inhibiting properties. The model is then used in drug discovery and development to predict the activity of compounds, for example, to evaluate whether it is worthwhile to synthesize them (i.e., are they predicted to have a high interaction with a given enzyme?). From a QSRR model the retention of a given compound on a given chromatographic system is predicted.
Many different modelling techniques can be used to build the model. Both linear and non-linear techniques are applied, such as multiple linear regression (MLR), partial least squares (PLS) and its variants, principal component regression (PCR), artificial neural networks (ANN), support vector machines (SVM) and multivariate adaptive regression splines (MARS).
The X-matrix usually contains many variables, mostly much more than the number of samples measured. The purpose in modelling is to build the simplest model with the best compromise between model fit and predictive properties. The modelling techniques thus require either a selection of informative variables from X or the initial X is reduced to a smaller matrix by calculating new variables, the so-called latent variables (e.g., the principal components or the PLS factors). Thus in many cases (e.g., when using MLR) a feature or variable selection is explicitly needed. The feature selection allows the most important variables from the many measured or calculated to be used, building simpler models and decreasing the risk of building an overfitting model.
An alternative approach is variable reduction, which means applying an approach to reduce the number of remaining variables by eliminating the least informative (2).
In feature selection, a learning algorithm is faced with the selection of a relevant subset of variables. Many feature selection techniques have been extensively used in the literature. Well-known techniques are genetic algorithms, forward selection, backward elimination and stepwise (selection and elimination), of which the latter three are usually combined with MLR. More recently, swarm intelligence optimizations, such as ant colony optimization and partial swarm optimization, which are selection techniques based on the behaviour of animals and insects, have been applied in, for example, QSAR studies. More information on the different feature selection techniques and their application in QSAR studies can be found in (3).
Three major categories of feature selection techniques can be distinguished: the filter, wrapper and hybrid methods (Figure 1). Filter methods perform an unsupervised feature selection. They do not use any information from the response y. The selection is based on a specific criterion, which is typically based on information content or inter-variable correlations in X. The wrapper methods are supervised feature selection methods, which use an objective function based on an optimization criterion to select the variables. The wrapper methods are computationally more expensive than the filter ones, but their generalization performance is better. The hybrid methods apply both above approaches. Usually in a first step a filter method is used to reduce the number of variables, followed by a wrapper method.
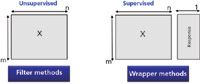
Figure 1: Feature selection: filter and wrapper methods.
Filter methods are often used in a first step to reduce the dimensionality of a data set. Constant, or nearly constant, variables, for example, are removed because of their lack of information. The filter methods can be divided in several types based on the criterion applied:
1. Dependency methods (e.g., correlation coefficient used as criterion),
2. Distance methods (e.g., Euclidean distance),
3. Information methods (e.g., entropy),and
4. Consistency methods.
Many more approaches are described in the literature (3).
When variables are removed based on their correlation, for example, then the two features that are most correlated are indicated and one can be randomly removed. Alternatively, in a better approach, the feature having the highest correlation with the dependent variable (y) is retained.
Wrapper methods use the information from both independent and dependent variables for feature selection. Several approaches are situated in this category and their number is increasing continuously. Forward selection, backward elimination and stepwise (selection and elimination), linked to MLR, are popular and well-known feature selection techniques.
In forward selection, the variable selected first has the highest correlation with the response y. Iteratively, one-variable-at-the-time is added to the model until all variables are added or the regression coefficient of the last entered is non-significant. In contrast, backward elimination starts with all variables in the model and then eliminates the least important one-by-one. The selection is finished when all remaining variables are significant. The stepwise procedure uses both forward selection and backward elimination. A variable that initially has been entered may in a later stage be eliminated because it became non-significant after the selection of other variables.
A genetic algorithm (GA) is another of the most popular feature selection techniques. It is a technique based on the natural evolution principles of Darwin. Features play the role of genes and a set of features represents a chromosome. Each object of a population is described as a chromosome with binary values, zeros and ones, representing the non-selected and selected variables, respectively.
The first generation is selected randomly but to come to a final selection of variables some parameters, such as the population size, generation gap, cross-over rate and mutation rate, need to tuned. Cross-over is an operation in which a pair of chromosomes partly exchanges information, while mutation is a genetic operation in which changes from one to zero and vice versa are introduced for a small fraction of the genes. The latter is done to create genetic diversity from one population of chromosomes to the next.
Many more feature selection techniques, among which the above mentioned swarm intelligence methods, have been applied in the literature. An overview can be found in (3).
The filter and wrapper methods are mainly applied to reduce the dimensionality of QSAR/QSRR data sets (i.e., when descriptors are used as X and MLR as the modelling technique). In multivariate calibration, when using spectra or chromatograms as X, variable reduction approaches are used to reduce the dimensionality of X. In these approaches the least informative or non-significant features are eliminated. This can, for example, be done in an uninformative variable elimination (UVE) approach, usually linked to PLS (UVE-PLS), or based on predictor-variable properties (2).
Uninformative variable elimination PLS (UVE-PLS) uses PLS to model, but the variables that are not more informative than noisy variables are removed from the data (4). The removal is achieved by initially adding artificially generated noise variables (and thus uninformative) to the original data. Then the experimental variables that are not more important than those added are eliminated. A reliability criterion for each original and each added random variable is determined to retain only the experimental variables for which the value of the reliability criterion is larger than the values obtained for the random variables.
This reliability criterion includes magnitude and uncertainty of the regression coefficients. Finally, the criterion values for the original variables are compared with those obtained for the artificial random variables. The original variables, for which the absolute criterion values are smaller than for the random ones (or for a selected fraction, for example, 99%, of the random variables), are considered uninformative and deleted. Their contribution to a regression model is thought to be small or negligible. Finally classical PLS is applied on the remaining variables, mostly resulting in a less complex model compared with classical PLS. UVE-PLS models usually have an improved predictive power compared with PLS ones (4).
Feature reduction approaches, where variables are sequentially eliminated, can be based on so-called predictor-variable properties. Examples of these properties are the absolute values of the PLS regression coefficients, the significance of the PLS regression coefficients, the norm of the loading weight vector, the variable importance in the projection (VIP), the selectivity ratio, the squared correlation coefficient of a predictor variable with the response y or combinations of them (2). The variable with the smallest value for the considered property is eliminated, the model rebuilt and the property values reconsidered to eliminate the next variable etc. Again the goal is to come to simpler models with better predictive properties.
J.P.M. Andries is a retired lecturer at the Avans Hogeschool, Breda, The Netherlands, and is working on a Ph.D. thesis about variable reduction in multivariate calibration.
M. Goodarzi is a Ph.D. student at the Vrije Universiteit Brussel, Belgium. He is working on a thesis about variable selection in QSAR/QSRR. Yvan Vander Heyden is a professor at the Vrije Universiteit Brussel, Belgium, Department of Analytical Chemistry and Pharmaceutical Technology, and heads a research group on chemometrics and separation science. He is also a member of LCGC Europe's editorial advisory board. Direct correspondence about this column should go to "Practical Data Handling", LCGC Europe, ,4A Bridgegate Pavillion, Chester Business Park, Wrexham Road, Chester CH4 9QH, UK, or e-mail the editor, Alasdair Matheson, at amatheson@advanstar.com
References
1. D.L. Massart et al., Handbook of chemometrics and qualimetrics: Part A, Elsevier, Amsterdam, (1997).
2. J.P.M. Andries, Y. Vander Heyden and L.M.C. Buydens; Analytica Chimica Acta, 705, 292–305 (2011).
3. M. Goodarzi, B. Dejaegher and Y. Vander Heyden; Feature selection methods in QSAR studies; in press (Journal AOAC International).
4. V. Centner et al., Anal. Chem., 68, 3851–3858 (1996).

Removing Double-Stranded RNA Impurities Using Chromatography
April 8th 2025Researchers from Agency for Science, Technology and Research in Singapore recently published a review article exploring how chromatography can be used to remove double-stranded RNA impurities during mRNA therapeutics production.
The Effect of Time and Tide On PFAS Concentrations in Estuaries
April 8th 2025Oliver Jones and Navneet Singh from RMIT University, Melbourne, Australia discuss a recent study they conducted to investigate the relationship between tidal cycles and PFAS concentrations in estuarine systems, and offer practical advice on the sample preparation and LC–MS/MS techniques they used to achieve the best results.











