Troubleshooting LC Separations of Biomolecules, Part 1: Background, and the Meaning of Inertness
How do bioinert and biocompatible liquid chromatography (LC) systems and columns improve separations of biomolecules? How do I know when these systems are required for my separation?
How do bioinert and biocompatible liquid chromatography (LC) systems and columns improve separations of biomolecules? How do I know when these systems are required for my separation?
We are witnessing tremendous growth in the life science and biopharmaceutical research areas, and separation scientists have risen to the myriad challenges that have presented themselves in these industries. Increasingly, analytical workflows are being designed with special measures taken to improve separation performance for biomolecules. Thus, terms like bioinert, biocompatible, biocolumn, and large molecule liquid chromatography LC (bio-LC) have arrived on the liquid chromatography scene. For this month’s instalment of “LC Troubleshooting” I’ve asked two my research collaborators, Jordy Hsiao and Greg Staples, to join me to address these topics from the point of view of troubleshooting. In this first instalment, we’ll dive into some of the background behind these terms and their relevance to bioseparations, and how examples of paying attention to specific characteristics of LC systems and separation conditions can have dramatic effects on the separation of biomolecules.
Dwight Stoll
Background
Several years ago, some of our own work focused on using hydrophilic interaction chromatography (HILIC) to separate biologically relevant small molecules. The preliminary results were quite exciting as we were able to achieve good separations of underivatized amino acids. However, we had difficulty detecting and obtaining good peak shapes for acidic metabolites containing phosphate groups or multiple carboxylate groups. These compounds are important because they are involved in many crucial cellular pathways (for example, the tricarboxylic acid cycle). At that time, we embarked on a journey to systematically study these effects, with the hope that a more detailed understanding of the observations would lead to better separations in the long run. It turned out to be a long trip, and the path is one that many analytical scientists have travelled. We began assembling other reports of deleterious interactions between biomolecules and high performance liquid chromatography (HPLC) systems, cataloging problems with phosphopeptides (1,2), phosphorylated glycans (3), monoclonal antibodies (mAbs) (4), and pharmaceutical compounds (5). From this survey, we observed that these problems were reported to occur on a variety of stationary phases in addition to HILIC, including reversed-phase, ion exchange, size-exclusion chromatography (SEC), and others. We were eventually able to develop robust, high performing separations of acidic metabolites. We hope some of the general lessons we’ve learned through these experiences can be leveraged by others to save significant time in tackling tricky biomolecule separations.
How to Spot Problems in Bioseparations
Poor performance for separations of biomolecules can manifest in a number of ways. Biomolecules as a class of analytes comprise a vast array of molecules that vary in terms of both physicochemical properties and size, and it’s important to note that chromatography problems can arise for species that are either small (such as metabolites, glycans, peptides), or large (such as proteins).
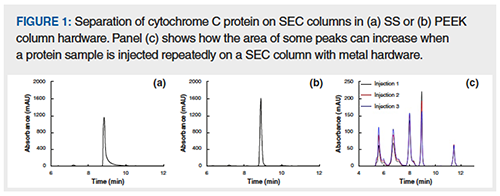
- Peak tailing: Often, analytes are eluted with undesirable peak tailing, which can make quantitation difficult. Sometimes these peak shapes gradually improve over the course of days or weeks, while other times their poor shapes can remain the same or continue to worsen. A good example of such peak tailing for a biomolecule is the separation of the protein cytochrome C using SEC. Figures 1(a) and 1(b) show such a separation on a column housed in stainless steel (SS) versus a column housed in polyether ether ketone (PEEK). The tailing factor of the protein on the SS column is significantly worse than on the PEEK column.
- Drifting peak area: Another situation that is commonly observed is an increase in peak area upon repeated injections of a sample, which can apply to individual or groups of analytes in the sample. An example of this is shown in Figure 1(c). The peak areas can eventually stabilize, but, in some cases, the stabilization can be very slow and require many injections. Sometimes, it’s impossible to be sure that the separation has stabilized at all.
- No observable elution: Perhaps most concerning are situations where an analyte is not detected at all because it is stuck somewhere inside the instrument between the sample vial and the detector.
We’ll address some of the countermeasures for the situations listed above in future instalments of “LC Troubleshooting”. In the meantime, you might be wondering about the root cause of these problems. There are unfortunately many culprits, but a common issue involves the interaction between biomolecules and metals in HPLC systems, the most common of which is the iron in stainless steel. There are several ways that biomolecules can interact with iron, in a physicochemical sense. For example, the phosphate groups in metabolites, glycans, and phosphopeptides can act as Lewis bases that tend to interact strongly with Lewis acids, such as iron. Moreover, molecules that contain multiple acidic functional groups (for example, malate, adenosine diphosphate) can bind metals with extraordinary affinities as a result of interaction geometries that are particularly favourable (for example, this is what makes the interaction between ethylenediaminetetraacetic acid [EDTA] and metals so strong). With proteins in particular, it is difficult to develop clear and robust rules about what will or will not interact strongly with metals because there is the potential for so many amino acid sequence-specific effects, as well as a strong dependence of the interaction on secondary and tertiary structure of the protein.
Bioinert to the Rescue!
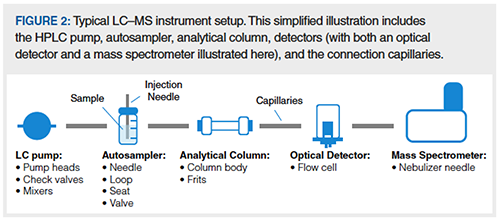
Let’s return to the terms we called out earlier, bioinert and biocompatible. These terms are used interchangeably in the literature, and in the context of biomolecule separations describe products that are designed to reduce problematic interactions with biomolecules. This is primarily accomplished by using alternatives to SS in the flow path of the separation. Some examples of materials that can be found in bioinert products are PEEK, alloys of titanium, ceramic, and MP35N (an alloy whose main components are Ni, Co, Cr, and Mo). Now that we are thinking of each of these different materials, it’s a good time to reflect on the many components of a UHPLC–mass spectrometry (MS) system, and consider how they relate to the interaction of biomolecules with metals. Figure 2 shows a block diagram of a typical LC–MS system. Metal-containing components can generally be divided into two categories: those that physically contact the sample and those that do not. The former includes things like the sample vial, the autosampler needle and loop, the HPLC column, any optical flow cells, transfer capillaries, and, in the case of MS, the nebulizer needle. The latter includes solvent bottles, pump heads, and mixers. It’s also worth noting that samples of biological origin may themselves contain metal ions, either inherently or by design.
Column Technologies for Biomolecules
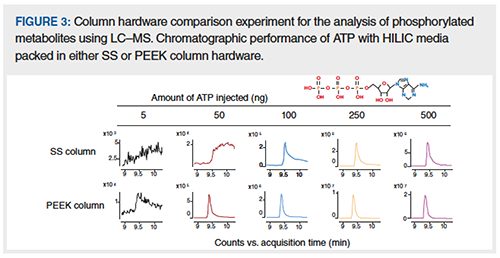
Analyte interaction with HPLC columns has been the subject of much attention from column manufacturers. Analytes can potentially interact with the walls of the column tubes, as well as the inlet and outlet frits that hold the stationary phase in place. Despite their small size, column frits can actually have surface areas in the same order of magnitude as the column walls. In some of our own work, we compared SS and PEEK HILIC columns for adenosine triphosphate (ATP) analysis. In the experiment shown in Figure 3, low amounts of ATP injected onto SS columns were not detectable. Only with larger injection amounts (above 250 ng) does the peak shape and intensity for ATP improve. This effect is mitigated using PEEK-lined columns, and more importantly, the sensitivity and peak shape improved significantly using PEEK hardware. Another similar comparison study has demonstrated better recovery and peak shape for phosphorylated N-glycans when using PEEK-lined column hardware (3). Alternatives to SS column hardware are available from column vendors, and comprise a variety of different bioinert materials in addition to PEEK (for example, titanium, glass).
Biocompatible HPLC Instrument Components
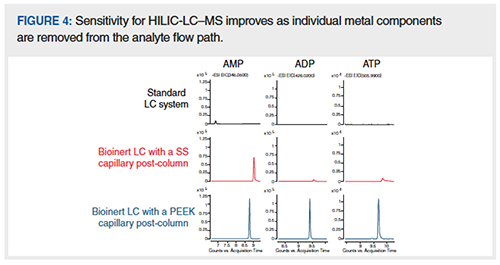
Let’s move on to the components of the HPLC–MS system that are relevant to metal–analyte interaction, keeping in mind the components in the flow path in Figure 2. It’s intuitive that there can be problems when the sample is in contact with a metal surface, but what about metal surfaces which the sample does not contact, like the pump heads? Indeed, these surfaces can be problematic when they leach (often variable) levels of metal ions into the mobile phase (6). This phenomenon can cause some serious complications, especially if leached metal ions accumulate on the column stationary phase, rendering the use of bioinert column hardware ineffective. Given that an HPLC–MS system has many potential sites of metal introduction or interaction, the effect on the performance of a biomolecule separation is cumulative. To illustrate this, consider the data in Figure 4, which examines separations of the metal sensitive analytes AMP, ADP, and ATP as markers for HILIC-LC–MS performance. We initially started with an LC system composed of a SS pump head, an autosampler with a SS injection needle, and SS capillaries connected to a quadrupole time-of-flight (QTOF) mass spectrometer (top row). Next, we changed the HPLC system to one with a titanium pump head, ceramic injection needle, and PEEK-lined connection capillaries, but with a single SS capillary post-column (middle row). In the last step, the postcolumn capillary was changed from SS to PEEK (bottom row). These results demonstrated that the signal intensities significantly increased as individual SS components were removed from the sample flow path. More notably, even a single SS capillary can negatively impact the detection limits for phosphorylated metabolites (compare middle and bottom rows).
Summary
In this first instalment of “LC Troubleshooting”, we’ve worked to highlight some of the problems you may encounter when developing a method for the analysis of biomolecules that are related to deleterious interactions between the analytes and the LC system. Such problems often show up in the form of lower than expected sensitivity, poor peak shape, and poor reproducibility. When working with analytes that have the potential to interact strongly with metals, it’s useful to know what to look for and how you can assess the overall performance of your system. In the best case, such separations should be developed using systems that limit or eliminate SS components. When this is not possible, there are other options available, including passivation and mobile phase additives, and we’ll discuss details of these approaches in a future instalment of “LC Troubleshooting”. Arming yourself with knowledge about the sources of metals in LC systems and the mechanism of interaction of biomolecules with these metals can be helpful in any troubleshooting you do, and facilitate the development of robust methods for accurate biomolecule determinations.
References
- A. Fleitz, E. Nieves, C. Madrid-Aliste, S.J. Fentress, L.D. Sibley, L.M. Weiss, R.H. Angeletti, and F.Y. Che, Anal. Chem.85(18), 8566−8576 (2013). doi: 10.1021/ac401691g.
- D. Winter, J. Seidler, Y. Ziv, Y. Shiloh, and W.D. Lehmann, J. Proteome Res. 8(1), 418−424 (2009). doi: 10.1021/pr800304n.
- K. Sandra, J. Vandenbussche, and P. Sandra, LCGC Europe31(10), 566–571 (2018).
- J.A. Anspach, S. Rao, and B. Rivera, LCGC North Am.36(6s), 24–29 (2018).
- M. De Pra, G. Greco, M.P. Krajewski, M.M. Martin, E. George, N. Bartsch, and F. Steiner, J. Chromatogr. A1611, (2020). doi: 10.1016/j.chroma.2019.460619.
- M.R. Euerby, C.M. Johnson, I.D. Rushin, and S. Tennekoon, J. Chromatogr. A705, 229−245 (1995).
Dwight R. Stoll is the editor of “LC Troubleshooting”. Stoll is a professor and co-chair of chemistry at Gustavus Adolphus College in St. Peter, Minnesota, USA. Direct correspondence to: LCGCedit@mmhgroup.com
Gregory Staples leads an R&D team focused on creating and developing separation, reagents, sample preparation, and analysis solutions for biomolecules at Agilent Technologies, in Santa Clara, California, USA.
Jordy Hsiao is an R&D scientist at Agilent Technologies, in Santa Clara.


.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)



