The Origin and Implications of Artifact Ions in Bioanalytical LC–MS
Liquid chromatography–mass spectrometry (LC–MS) with electrospray ionization (ESI) is a widely used bioanalytical technique with both qualitative and quantitative applications. Ions are created by electrically charging a stream of droplets from the LC system, which evaporate and leave ions that are transferred to the mass spectrometer. Ideally, these are only from the analyte, but background ions, such as metals, impurities and coeluted species, can react with analytes producing adducts, such as [M + Na]+, [M + K]+, and multimers (2M + H+, 3M + H+, and so forth). Although well known, the extent of adduct ion formation and the implications for quantitative analysis and analyte characterization by tandem MS (MS/MS) are not fully appreciated. We summarize the problem and identify areas that should be considered when developing or using electrospray LC–MS.
Modern liquid chromatography–mass spectrometry (LC–MS) is dependent on atmospheric pressure ionization (API) techniques, such as atmospheric pressure chemical ionization (APCI) and electrospray ionization (ESI). ESI is widely used for analyzing polar molecules in biological fluids and enables applications such as proteomics and metabolomics as well as analyses in drug development, toxicology, and doping control. It is compatible with a wide range of LC flow rates and usually produces protonated ([M+H]+) or deprotonated ([M-H]-) ions that can be dissociated to generate structurally informative fragments using collision-induced dissociation (CID). The combined technique is known as LC–tandem MS (MS/MS) because it uses two mass selection steps, one to select a precursor ion and the other to analyze the fragments following CID. Selectively monitoring fragment ions from a preselected precursor ion provides a sensitive quantitative method known as selected reaction monitoring (SRM) that is widely used in drug development and legal toxicology applications. Analysis of the fragment ions is used for compound identification, either by comparison with spectral libraries or de novo, preferably using a high-resolution analyzer that generates accurate mass values from which likely elemental compositions can be determined. The process has been automated in a technique known as data dependent analysis (DDA), where candidate precursor ions are determined from one mass analyzer scan, individually selected and fragmented, and the product ions analyzed. This technique is the basis of proteomics. With DDA, the selection window is generally 1 Da wide to increase the specificity, but it was recently realized that if the fragments are sufficiently specific (for example, because of high-resolution mass analysis), wider selection windows can be used. This process has become known as data independent analysis (DIA) and has several advantages, primarily that a wide range of precursor ions can be monitored continuously and that all compounds are fragmented, generating a reproducible permanent record of the sample. For quantitative DIA, as used in proteomics, data analysis is based on the chromatographic behavior of fragment ions known to be produced by the target compounds.
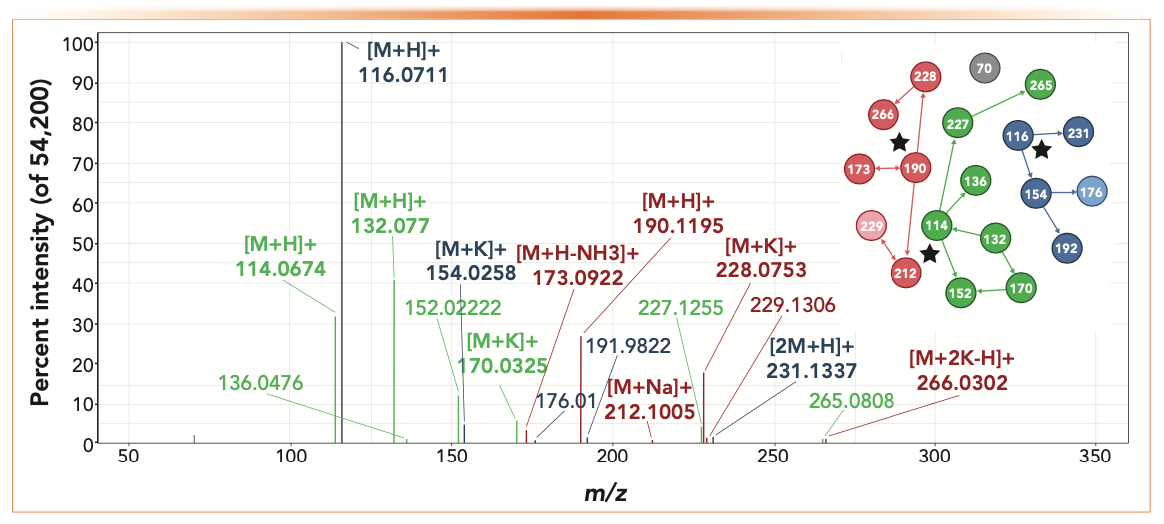
ESI ions are created by electrically charging a stream of droplets from the LC system, which evaporate and leave ions that are transferred to the mass spectrometer. Ideally, these only come from the analyte, but ions from involatile entities, such as metals and impurities present in the sample or from the instrument, can also react with analyte molecules and produce adducts, such as M + Na+, M + K+, and M + NH4+. Although these are well-known artifacts, the full extent and implication of these reactions has only been recently realized. For example, Mahieu and others (1) reported in 2017 that 25,000 LC–MS features could be reduced to 1000 and Broeckling (2) in 2016 noted that much of the signal measured in the spectra of known compounds could not be explained. In fact, Broeckling’s paper contained a spectrum of dimethylsuccinic acid (Figure s2 in that paper) where the [M + H]+ ion is weak, and the ions containing sodium and potassium are more intense. However, the bulk of the intensity is because of the multimers (2M and 3M) combined with potassium and also with other, unknown entities. We encountered similar phenomena while building spectral libraries by infusing standards (3) and developed software to recognize the more common species (4); however, like Broeckling, many species remained unidentified, which can be particularly challenging when several compounds are coeluted, as illustrated in Figure 1.
FIGURE 1: Annotated, background subtracted, deisotoped, and thresholded full scan spectrum of LC–MS analysis with coelution of L-proline (blue), homocitruline (red), and creatine (green), adapted from reference (4). The inset shows the networks of related ions with the assigned MH+ ion indicated by a star notation.
We subsequently developed an approach called multi-layered analysis (MLA) to focus on unidentified ions (Figure 2), which revealed multimers and adducts with calcium, aluminum, iron, and barium, and potentially magnesium, zinc, and molybdenum in our spectrum and calcium, aluminum, and iron in Broeckling’s. These metals apparently arise from parts-per-billion (ppb)-level trace elements in the solvents used (particularly formic acid) although some, like potassium, are present in biological fluids. Although sodium and aluminum are monoisotopic, the others have patterns that can aid interpretation, such as barium, but they can also overlap and produce more complex patterns (5).
FIGURE 2: Multi-layered analysis for succinic acid, adapted from reference (5).
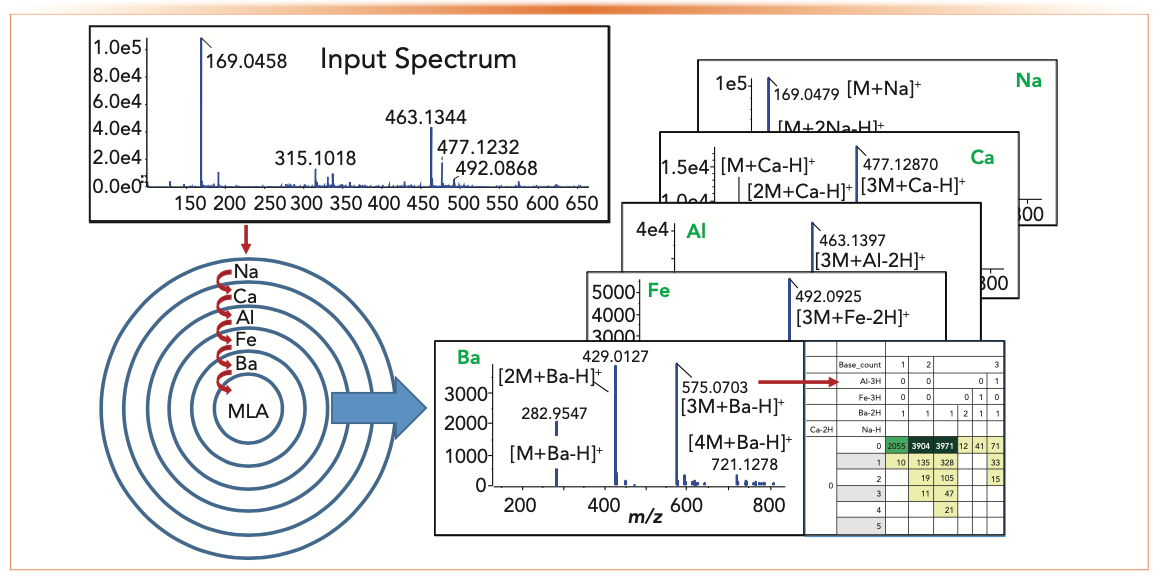
As part of this study, we calculated adduct masses using effective adduct mass (MA) which is equal to A – vH, where A is the mass of the adduct, v is its valency, and H is the mass of hydrogen, plus the number of protons required by the observed charge. Therefore, the basis of barium adducts with a valency of 2 is M+ (Ba – 2H) giving singly charged ions with m/z M + (Ba – 2H) + H+ and doubly charged ions at (M + (Ba–2H) + 2H+)/2, equivalent to (M + Ba2+)/2. Using this approach emphasized two important facts. First, elements with different masses and valency can produce adducts that are close in mass and can be resolved by their isotope patterns (for example, ions from potassium and calcium differ by 0.009 Da). Second, the same element in different oxidation states can also produce overlapping patterns; for example, Fe(II) and Fe(III) are shifted by the mass of hydrogen, which is 1.007825. Furthermore, adducts can involve combinations of elements (for example, M + Na + Al) and the addition of background and coeluted species, making complete interpretation difficult. We also note that sample preparation can also produce byproducts that complicate the analysis, but these are different compounds that may be separated, whereas adducts are ion source phenomena and have the same retention time as the analyte.
Implications
Regardless of the exact source and composition of adducts, their presence has important implications for LC–MS analyses, especially for the complex samples often encountered in bioanalysis.
Reproducibility
The formation of adducts and ions from reactions between analytes and background ions or between coeluted analytes is highly dependent on the sample matrix, the purity of the reagents and solvents, the cleanliness of the system, and the separation used. Thus, the type and extent of these species is liable to inter-instrument, inter-laboratory, and temporal variation, affecting the intensity of the observed ions, which would impact all of the areas discussed below and may confound long-term studies such as proteomics and metabolomics. The ability to transfer methods between laboratories will also be affected (for example, if the reagents used are from different sources). We speculate that this problem may be part of the reason for the anecdotal difficulty in transferring regulated clinical trial assays between pharmaceutical companies and contract research organizations (CROs).
Qualitative Analyses
As noted, adducts can dramatically increase the number of LC–MS features observed, which complicates interpretation of the results. We have noted that this complication is especially true in analyzing high-molecular-weight compounds, such as peptides and glycopeptides, because these have extended 13C isotope patterns and often form multiply charged ions that reduce m/z isotope spacing. As a result, adducts can produce complex overlapping isotope patterns that result from simple mass shifts for adducts that have no isotopes (sodium, aluminum, cobalt, and manganese) or altered patterns if the adduct itself has isotopes or different oxidation states (iron, molybdenum, and cadmium). In some cases, real modifications may be confused with adducts of similar mass, and if these are coeluted, the observed mass may not correspond to either. For example, the effective adduct mass of cobalt is 56.91754, and an acetamide derivative differs by 57.021464, and although the m/z difference of 0.104 is usually resolved, it decreases with increasing charge and the species may eventually merge, giving a peak at 56.96950 (assuming equal intensity), which differs by 0.053 from both species.
Quantitative Analyses
Because adducts arise from real analyte molecules, they may reduce the intensity of the expected ions and cause inaccuracy, which could be particularly dangerous in targeted assays, such as SRM and multiple reaction monitoring (MRM), because the full spectrum is not usually recorded and the adduct ions may be undetected. Furthermore, the introduced variability will cause the error to change over time. We also speculate that the lengthy development required for many regulated pharmaceutical assays may, in part, arise from the unrecognized need to control adduct formation to obtain the necessary linear dynamic range.
DDA and DIA
DDA is a fully automated process controlled by the instrument manufacturer’s software and, to our knowledge, does not consider adduct formation. Thus, adduct ions are as likely to be selected, especially if they are intense or if deep selection is used, resulting in low-quality spectra that are difficult to interpret because adduct ions often fragment poorly. We further speculate that this may be part of the reason that many spectra obtained in proteomic analyses are unassigned. However, we reported that alternative fragmentation techniques can produce spectra from adduct ions with complementary information that aids interpretation (6).
In DIA techniques, such as sequential widowed acquisition of all theoretical fragment ion mass spectra (SWATH) (7), all ions are fragmented and adduct ions may be useful. For example, in a simple metabolomics study (8), we noted a change in an LC–MS feature that was identified as a potassium adduct and, because this was SWATH data, we followed the fragmentation chain and identified the glucuronide derivative of a known metabolite. Further, SWATH allows comparison of MS2 data to help identify related species.
Conclusion
Although well known, the extent and implication of adduct ion formation may not have been fully appreciated. As discussed, adducts often cause additional complexity, may reduce sensitivity in targeted analyses, and introduce uncontrolled variability that challenges reproducibility. However, they may also provide opportunities for alternate fragmentation methods that could be exploited by instrument manufacturers. They can also help confirm the molecular weight, especially if the [M+H]+ ions are absent and may reveal structural information if formation is dependent on the structure.
Regardless, because moieties potentially causing adducts are unavoidable in biofluids and reagents, it is important that the possibility of adduct formation (especially with unusual or unexpected elements) is considered and treated appropriately or that precautions are taken to avoid undesirable consequences.
References
(1) N.G. Mahieu and G.J. Patti, Anal. Chem. 89(19), 10397–10406 (2017). DOI: 10.1021/acs.analchem.7b02380.
(2) C.D. Broeckling, A. Ganna, M. Layer, K. Brown, B. Sutton, E. Ingelsson, G. Peers, and J.E. Prenni, Anal. Chem. 88(18), 9226–9234 (2016). DOI: 10.1021/acs. analchem.6b02479.
(3) T. Bruderer, E. Varesio, A.O. Hidasi, E. Duchoslav, L. Burton, R. Bonner, and G. Hopfgartner, Anal. Bioanal Chem. 410(7), 1873–1884 (2018). DOI: 10.1007/s00216-018-0860-x.
(4) T. Stricker, R. Bonner, F. Lisacek, and G. Hopfgartner, Anal. Bioanal Chem. 413(2), 503–517 (2021). DOI: 10.1007/s00216-020-03019-3.
(5) R. Bonner and G. Hopfgartner, Anal. Chim. Acta. 1193, 339317 (2022). DOI: 10.1016/j.aca.2021.339317.
(6) A.O. Ducati, D. Ruskic, P. Sosnowski, T. Baba, R. Bonner, and G. Hopfgartner, Anal. Chim. Acta. 1150, 338207 (2021). DOI: 10.1016/j.aca.2021.338207.
(7) L.C. Gillet, P. Navarro, S. Tate, H. Rost, N. Selevsek, L. Reiter, R. Bonner, and R. Aebersold, Mol. Cell Proteomics 11(6), 16717 (2012). DOI: 10.1074/mcp.O111.016717.
(8) R. Bonner and G. Hopfgartner, Bioanalysis 8(16), 1735–1750 (2016). DOI: 10.4155/bio-2016-0141.
Ron Bonner runs Ron Bonner Consulting in Newmarket, Ontario, Canada. Gérard Hopfgartner is with the Department of Inorganic and Analytical Chemistry at the University of Geneva, in Geneva, Switzerland. Direct correspondence to: gerard.hopfgartner@unige.ch
New Method Explored for the Detection of CECs in Crops Irrigated with Contaminated Water
April 30th 2025This new study presents a validated QuEChERS–LC-MS/MS method for detecting eight persistent, mobile, and toxic substances in escarole, tomatoes, and tomato leaves irrigated with contaminated water.
Accelerating Monoclonal Antibody Quality Control: The Role of LC–MS in Upstream Bioprocessing
This study highlights the promising potential of LC–MS as a powerful tool for mAb quality control within the context of upstream processing.
University of Tasmania Researchers Explore Haloacetic Acid Determiniation in Water with capLC–MS
April 29th 2025Haloacetic acid detection has become important when analyzing drinking and swimming pool water. University of Tasmania researchers have begun applying capillary liquid chromatography as a means of detecting these substances.
Prioritizing Non-Target Screening in LC–HRMS Environmental Sample Analysis
April 28th 2025When analyzing samples using liquid chromatography–high-resolution mass spectrometry, there are various ways the processes can be improved. Researchers created new methods for prioritizing these strategies.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)











