Stoichiometry of Intrinsically-Disordered Protein Complexes
The Application Notebook
Intrinsically-disordered proteins cannot be analyzed by SEC column calibration. SEC-MALS provides accurate molecular masses of these proteins and of their complexes with other proteins.
Intrinsically-disordered proteins cannot be analyzed by SEC column calibration. SEC-MALS provides accurate molecular masses of these proteins and of their complexes with other proteins.
The S100 family is a group of dimeric, calcium-binding proteins which have been found to be overexpressed in several types of cancer. S100 proteins bind to the C-terminal region (293-393) of p53, the most critical tumour suppressor involved in apoptosis, cell cycle arrest, and DNA repair. Since S100 is dimeric and the C-terminus of p53 encompasses a tetramerization domain, the potential range of complexes incorporating p53 and S100 is large.
The p53 variants have a large fraction of intrinsically disordered structure and do not elute according to globular protein standards in size-exclusion chromatography (SEC). However, multi-angle light scattering (MALS) determines molecular mass from first principles. SEC-MALS determines the molecular weight of the complexes independently of protein standards. We utilized SEC-MALS to investigate how S100 proteins bind to p53 in its different oligomeric states, using monomeric (L344P) and dimeric (L344A) p53 mutants.
Experimental Conditions
The experimental system consisted of an HPLC and SEC column plumbed in series with a DAWN MALS detector (Wyatt Technology) and an Optilab differential refractive index (dRI) detector (Wyatt Technology).
The p53 mutants and S100 proteins were first characterized separately by SEC-MALS to determine their molecular masses and homogeneity. They were then incubated at various stoichiometric ratios-4:1, 2:1, 1:1, 1:2, and 1:4-and analyzed on the SECâMALS system.
Results
Molar mass results of the individual p53 proteins are overlaid with the light scattering (solid lines) and dRI chromatograms (dotted lines) in Figure 1. The molar masses correspond to a monomer of 11.2 kDa for the L344P variant, a dimer for the L344A variant, and a tetramer for p53 wild-type.
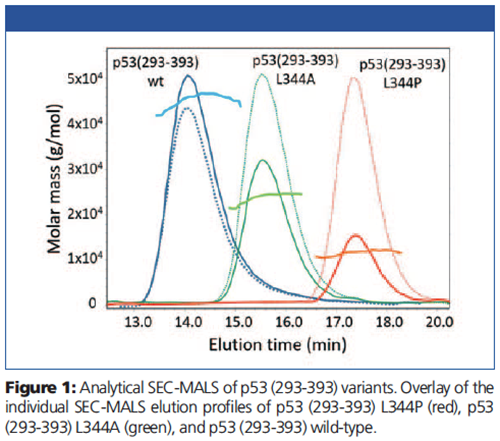
Chromatograms and molar masses for the pre-incubated mixtures are presented in Figure 2. Monomeric p53 (293-393) L344P (11.2 kDa) and dimeric S100B (21.4 kDa) both eluted with a retention time of ~17.5 min.
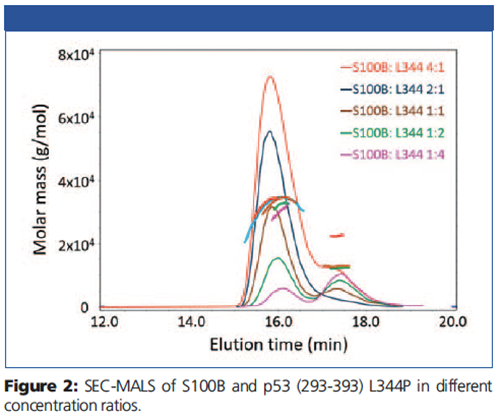
The complex molar mass of ~32 kDa at ~16 min is independent of the relative ratio of the two proteins, indicating that one dimer of S100B binds only one monomer of p53. No complexes consisting of a dimer of S100B and two monomers of p53 (~43 kDa) could be detected. In addition, S100B was able to disrupt the dimeric p53 (293-393) L344A mutant and form a complex with the monomer.
Interestingly, SEC-MALS revealed that S100B was also able to form a stable complex with tetrameric p53 (293-393) consisting of 4 dimers of S100B bound to a tetramer of p53.
Conclusions
SEC-MALS helped understand how S100 proteins influence the oligomerization of p53. Based on these results and in vivo studies, we established a binding model where S100 proteins can activate tetrameric p53 but inhibit p53 activity binding to the monomer by shifting the tetramerization equilibrium. SEC-MALS is a valuable tool in structural biology for studying the stoichiometry and formation of many types of protein-protein complexes.

Wyatt Technology
6330 Hollister Avenue, Santa Barbara, California 93117, USA
Tel.: +1 (805) 681 9009
Website: www.wyatt.com

MALDI Guided SpatialOMx® Uncovers Proteomic Profiles in Tumor Subpopulations of Breast Cancer
September 1st 2020The timsTOF fleX system bridges a current gap by providing MALDI Imaging and in-depth proteomics analysis in just one instrument. The instrument offers all benefits of a timsTOF Pro for time-efficient and sensitive proteomics, combined with a high-resolution MALDI source and stage. Using PASEF technology, it is possible to retrieve high protein ID rates with small sample amounts. Here we present the new SpatialOMx® workflow to identify distinct proteomic profiles for different tumor subpopulations in breast cancer as an example for this powerful approach.





