Progress in Peak Processing
Special Issues
The advantages and limitations of several recently introduced mathematical procedures for enhancing peak resolution in liquid chromatography (LC) are described. Despite advanced separation technologies and extensive knowledge in method development, peak overlap is still commonly observed. This article gives a brief overview of the advantages and limitations of recently introduced mathematical procedures for enhancing resolution.
Despite advanced separation technologies and extensive knowledge in method development, peak overlap is still commonly observed. Peak integration becomes more challenging as chromatographic resolution decreases, especially with asymmetric peaks. Post-acquisition signal processing, well established in optical spectroscopy, and nuclear magnetic resonance (NMR), is now being used in liquid chromatography (LC). Mathematical operations can be applied on raw chromatographic data to enhance resolution of overlapping peaks and reduce peak widths. These techniques can maintain original area information needed for quantitation after some modifications. This article gives a brief overview of the advantages and limitations of recently introduced mathematical procedures, such as the Fourier deconvolution of extracolumn effects, iterative curve fitting, multivariate curve resolution, modified power law, and use of first and second derivatives in enhancing resolution. High-throughput analyses in gas chromatography (GC), LC, and supercritical fluid chromatography (SFC) could benefit from these simple and effective approaches in many challenging separations applications.
Some analytical chemists often wonder: What is the future direction of separation science? One school of thought holds that this field is mature, and not much remains to be done. Spectroscopy went through a similar phase a few decades ago, but the introduction of digital signal processing revolutionized the whole field of molecular spectroscopy and nuclear magnetic resonance (NMR) spectroscopy. It is impossible to imagine any modern infrared (IR) or NMR spectrum that has not undergone a Fourier transform or other mathematical manipulations. Separation scientists have been quite hesitant to adapt mathematical techniques to enhance peak resolution, but perhaps we can extract more from less, even if the physical separation is not fully developed. The purpose of analytical separations (for example, chromatography or electrophoresis) is to obtain useful information. This can be qualitative or quantitative in nature. Things that enhance the speed of the process and the accuracy of the information are highly desirable.
Advances in chromatography have led to highly efficient separations, and we are finally beginning to grasp the science behind high-efficiency columns (1–3). At best, randomly packed beds consisting of nonporous, superficially porous, and fully porous particles can produce reduced plate heights h (equal to the theoretical plate height divided by the particle diameter, H/dp) as low as 0.5, 0.7, and 0.9, respectively (4), whereas in practice we are currently halfway there. Davis and Giddings, on the basis of statistical theory of overlap, predicted that a multicomponent chromatogram should be roughly 95% empty, in order to provide a 90% probability that a given analyte of interest will appear as an isolated peak (5). Even with modern high efficiency separations, there are cases where one or two critical pairs have resolution problems; for example, deuterated versus nondeuterated molecules, enantiomers, or cases where there are large number of peaks. More often, in enantiomeric separations, the entire separation window is empty, and yet the enantiomers have poor resolution. Usually, there is an ambiguity in the integration of overlapped chromatographic peaks when using routine drop perpendicular skimming methods. Thus, the development and use of a method that suitably separates all the components necessary for quantitation (usually with the aim of a baseline separation, resolution = 1.5) commonly becomes the bottleneck of chromatographic analysis in research work, as well as in the pharmaceutical industry. What if, with a click of a button, resolution were instantaneously improved, and there were no need to go through the arduous process of method development (switching stationary phases or mobile phases)?
The primary concern is: Can we mathematically improve chromatographic resolution, while maintaining critical peak information necessary for quantitation? It would also be preferred if the protocol were simple and straightforward. In this article, the fundamental ideas that govern new signal processing protocols, including deconvolution, are given for Fourier transformation (6,7), iterative curve fitting and multivariate curve resolution (8–10), power laws (11,12), and derivatives (13). These are shown in Table I. They fall under three general categories: 1) elimination of extracolumn band broadening, 2) extracting peak areas by curve fitting, and 3) directly enhancing resolution by reducing peak widths. The following sections describe these strategies with their advantages and limitations, per the maxim that when we gain something, in turn we can lose something else. These resolution enhancement strategies generally require ubiquitous software (such as Microsoft Excel), single channel data, and will surely be implemented into chromatography data software in the future. Once fully automated, their true power will be most apparent in ultrafast (<1 min), and hyperfast (<1 s) liquid chromatography (LC) and high peak capacity separations.
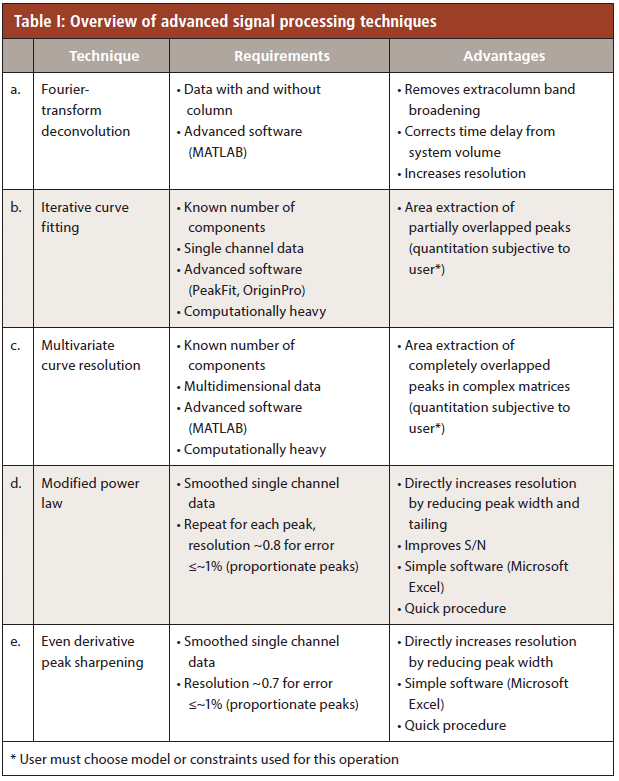
Deconvolution of Extracolumn Effects by Fourier Transformation (FT)
A chromatograph that does not contribute to band broadening has yet to be invented. The recorded signal from the instrument is convoluted with broadening by the injector, connection tubings, and the detector design. Deconvoluting this effect would remove these extracolumn effects from the chromatogram. Resolution would also increase if the separation were compromised by the hardware and software. FT deconvolution was first described in the early 1980s (7). Recent work evaluated the band broadening elimination by FT deconvolution on modern ultrahigh-pressure liquid chromatography (UHPLC) systems and narrow-bore columns, as shown in Figure 1 (6). The protocol for FT deconvolution is a three-step process. First, a chromatogram must be collected with and without the column (Figure 1a). Then, both chromatograms are converted to the frequency domain by Fourier transformation (Figure 1b). Next, the frequency transformed data from the chromatogram with the column are divided by the frequency transformed data collected without the column. The resulting quotient is converted back to the time domain by the inverse Fourier transform (6). This yields a chromatogram that is free of extracolumn band broadening effects (Figure 1c). There is a shift in peak retention time resulting from the time needed for the injected analyte to reach the detector without the column; that is, the system volume effect is also corrected. Baseline noise increases as a result of division in the frequency domain, because division by very small numbers as well as oscillations are seen. However, these can easily be decreased by digital smoothing or cutting off all high frequency noise (ωc). Fourier transform deconvolution has also been applied while working with 1-cm columns at extremely high flow rates (14).
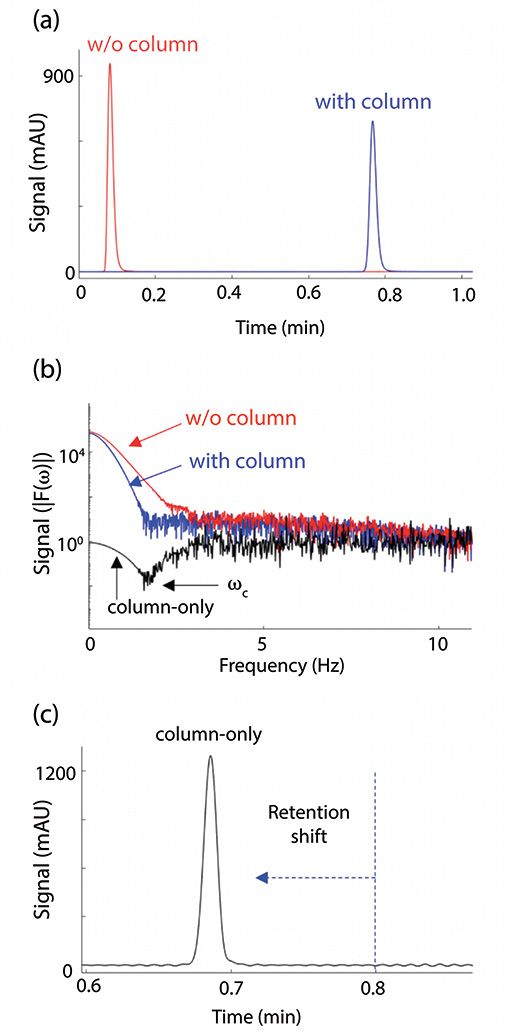
Figure 1: Removal of extracolumn band broadening effects by Fourier-transform deconvolution (6). The collection of a chromatogram with and without the column is shown in (a). Then, (b) each dataset is converted to the frequency domain. Next, (c) they are divided, with the result shown as “column-only”. This is converted back to the time domain. The retention time of the chromatographic peak has also shifted, accounting for the system volume. (Figures in MATLAB provided by Y. Vanderheyden)
Peak Area Extraction by Iterative Curve Fitting
Iterative curve fitting is a versatile approach for extracting peak areas from partially overlapping peaks, especially when multiple components are overlapping to some extent. The chromatogram, containing time and single-channel signal, is exported into a curve-fitting software; see for example Table I, which considers the entire chromatogram as a sum of exponentially modified peaks. It is assumed that a single peak represents a pure component. The number of components (peaks) is proposed by the user, and then the chromatogram is fitted according to the chosen peak model by method of minimization of residuals. There are several peak functions, but for LC an exponentially decaying tail is usually observed. The most useful model for these purposes has been determined to be the bidirectional exponentially modified Gaussian (BI-EMG), which is a Gaussian function with a one-sided exponentially decaying tail or front as a function of time (15). For simple chromatograms, one can conveniently obtain a fit with a coefficient of determination (R2) close to 1 (if R2 = 1, then it is a perfect fit). This is a trial-and-error approach, where the user continues to adjust the initial parameters of the model, iteratively improving the fit until it is considered acceptable. Caution should be exercised that an iterative curve fitting procedure may yield several mathematically correct answers. Similarly, it is ambiguous to fit several peaks under a single peak, which is mathematically possible, but it will not disclose the reality of the underlying peaks.
Once a suitable fit is determined for the separation, a baseline must be established to extract each underlying peak area. In most cases, a simple linear baseline is sufficient. However, in gradient elution or multidimensional separations, a nonlinear baseline could be utilized by choosing it from the software. The use of iterative curve fitting to extract peak areas from overlapping peaks is illustrated in Figure 2. A simulated separation of seven peaks in under a minute is shown. There are two sets of overlapping segments with differing degrees of tailing and efficiencies. Since it was simulated, the true area of each peak was known. The exact peak areas of peaks 1 to 7 were 4, 3, 6, 8, 5, 10, and 9 area units in the absence of noise, respectively. Using the BI-EMG model, this separation was fitted with an R2 of 0.9996. After this mathematical fitting, peak areas can be extracted, as well as other peak information, including efficiency, tailing factors, peak height, zeroth, first, second, and statistical moments. In this case, the extracted areas are in order of peaks 1–7 are: 3.99, 2.99, 5.99, 7.99, 5.01, 9.98, and 9.02, respectively, with an excellent match of theoretical areas in the presence of random noise. Overall, curve-fitting procedures are powerful for extracting peak areas when it is clear that there are no hidden peaks under the peak of interest. Choosing a pure Gaussian peak is only a limiting case because real peaks often have a "tail" or "front" better described by an EMG function.
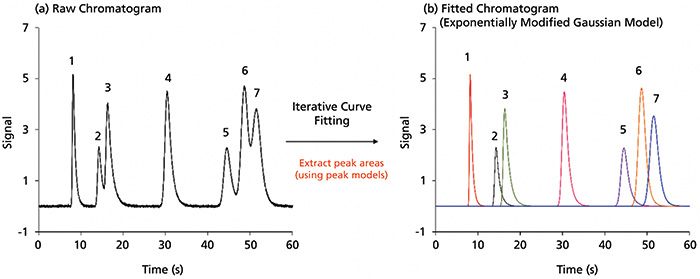
Figure 2: Iterative curve fitting of seven simulated peaks with different peak heights, areas, and shape. (a) The raw chromatogram obtained from the simulation are shown. (b) After fitting, using a bidirectional exponentially modified Gaussian model and a linear baseline, each peak area can be extracted. Customization can be made to the constraints, which improves the fit and allows the user to fit any peak shape.
Model-Free Approaches for Peak Information Extraction
Various powerful methods exist as well as iterative curve fitting for extracting peak information, even when the peaks overlap completely, where an iterative curve fitting method mentioned above will fail (16). Unlike iterative curve fitting, these methods require multidimensional data; that is, various signals are acquired at the same time. Second, these signals must be specific to the molecule of interest. For example, a photodiode array generates an entire spectrum of a given component; similarly, mass spectrometry (MS) generates an analyte-specific signal. Third, in order to identify multiple peaks in a completely coeluted peak envelope, the key requirements are that the compounds that are coeluted must be known, and their pure spectra must be present in the software library. The latest example is that of the vacuum ultraviolet (VUV) GC detector. The mathematical technique is termed as linear combination of weighted reference spectra . The VUV software can extract complete peak information of coeluting compounds if the spectra of the coeluted compounds are known and they are sufficiently distinct. The observed spectrum at each data point is treated as the sum of pure spectra for the coeluted compounds following equation 1:

where A1 and A2 are the pure absorbance spectra of each component, and f1 and f2 are corresponding scaling factors. These scaling factors are determined by linear regression by minimization of residuals. The fit coefficients f1 and f2 plotted over the time region of a coelution event represent chromatographic signals for each of the coeluting compounds. Measured VUV absorbance spectra can be converted into chromatographic signals using spectral filters (16).
Another tool that can estimate underlying elution and spectral profiles for a chromatogram is multivariate curve resolution–alternating least squares (MCR-ALS), even in the case of complete overlap of peaks (Rs = 0). The main requirement from a chromatographic point of view is to collect data from multiple channels, like the case of VUV. The availability of photodiode array detectors in high performance liquid chromatography (HPLC) systems has made this procedure convenient, because it allows the construction of a multidimensional data matrix. The goal of MCR-ALS is to decompose the observed data matrix (D) of a chromatogram into elution (C) and pure spectral profiles (ST) that optimally fit the data matrix as shown in equation 2. E is the experimental error in the estimated convergence:
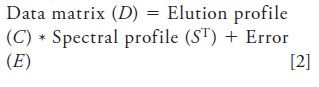
MCR-ALS requires an initial estimation of pure spectral profiles (ST). Perhaps the fastest way to get the initial estimate is if the components are known, and a pure spectrum is available for each component. If the components and their pure spectral profiles are not available, then the most common way is to estimate the concentration profiles using evolving factor analysis (EFA) (17), or simple-to-use interactive self modeling mixture analysis (SIMPLISMA). The details of EFA can be found in the seminal work by Maeder (17), and in examples in previous LCGC reviews on MCR-ALS (18) for peak purity analysis. Since MCR-ALS and the linear combination of weighted reference spectra approach used in VUV requires an initial estimate of concentration or spectral profile, enantiomers might be more difficult to differentiate, especially if there is no separation because their ultraviolet-visible (UV–vis) absorbance and their MS spectra would be identical. Similarly, universal response detectors cannot be used with MCR-ALS, which essentially eliminates all data from flame ionization detectors (FID), thermal conductivity detectors (TCD), barrier discharge ionization detectors (BID), conductivity detectors, and refractive index (RI) detectors. However, MCR-ALS is not limited to UV–vis or MS. In addition, this procedure is subjective to the user, because the constraints can be inappropriately chosen, and lead to unrealistic peak shapes. Most MCR methods use non-negativity and unimodality, but other various constraints, such as closure, trilinearity, selectivity, and other shape constraints, make MCR the most sophisticated technique among all described herein. When multiple peaks are determined under a similar curve, computation is more difficult, and can increase post-processing time. Some commercial spectroscopy software has already implemented MCR-ALS but, to our knowledge, most chromatography data software has not, except for one type (19).
Direct Resolution Enhancement by Power Law
Unlike MCR-ALS, the power law approach is a single-channel method, and it can be applied on any detectors not amenable to MCR-ALS. The power law directly increases chromatographic resolution (Rs) of overlapping peaks to baseline separation (Rs = 1.5), so they are easier and more accurately visualized and integrated (11,12). The fundamental principle of a recently proposed power law is that raising a given output signal to a power, n (where n is an integer >1), increases the signal magnitude if it is >1, or decreases the signal magnitude if it is <1 (11). The power law (or power transform) reduces tailing and noise, maintains retention time, and increases resolution between overlapping chromatographic peaks. Already, a simpler version of power law is integrated in some software (20), where collected chromatographic signal data can be raised to a power (max of n = 3), and then integrated normally. However, the simple law is not suitable for quantitation, because the relative area of exponentially enhanced peaks has changed after the mathematical operations relative to the original peaks (12). As a result, a modified power law approach was introduced in 2019, which maintained peak area integrity and offered all the benefits of a simple power law (11).
The modified power law relies on this fundamental characteristic by normalizing the peak of interest's maximum to a value of 1 (and the rest of the chromatogram accordingly), before raising the chromatographic signal to a power that provides the desired resolution. The chromatographic data can be exported to Excel and the peak area quantitated with an external method either in Excel, or by numerical integration. It is desirable to smooth the raw data and correct the baseline if a drifting baseline resulting from a gradient method has emerged. Each peak in a critical pair is first normalized to unit height followed by raising the chosen peak signal to a desired power. It is recommended to have Rs ≥ 0.8. The area recovery is described below in equation 3.
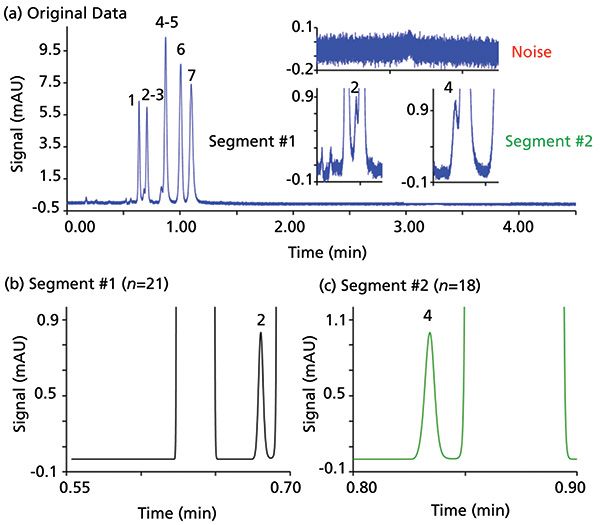
Figure 3: Directly increasing resolution of two overlapping pairs by modified power law. (a) The original separation data of hormones (in order of elution) is shown: 17α-ethynylestadiol, estrone, estriol, estradiol, androstadienone (and rosta-4,16-dien-3-one), progesterone, and testosterone. See reference 4 for chromatographic information. (b) and (c) show each overlapping pair baseline separated of each segment; segment 1 with a power (n) of 21 and segment 2 with a power (n) of 18. The area of peaks 2 and 4 can be recovered using equation 3. Adapted with permission from reference 20.
To visualize this method, an example from a recent article is shown in Figure 3, where two critical overlapping pairs are present and identified as segment 1 and segment 2 (20). Noise is high, and all chromatographic peaks are tailing, making integration difficult (Figure 3a). After applying powers in each segment (Figures 3b and 3c), peak widths are reduced, and signal-to-noise (S/N) is significantly enhanced. After raising these segments to powers, it is much easier to integrate, and the original peak area can be back-calculated using equation 3 where n = the power used to get baseline resolution:
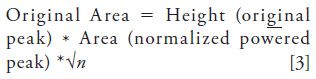
Questions that remain are: How is the correct power chosen, and how much error is there?
Originally, each pair had different magnitudes of overlap (more or less resolution) so different powers were needed to get a baseline resolution (Rs = 1.5). Choosing what power of n to use is somewhat arbitrary; that is, two overlapping peaks might be baseline separated by using a power of 3, but if a power of 10 were used one would still get higher resolution. Where do we stop? Since the chosen n is limitless (limit towards infinity), very large powers could be chosen. However, if such large powers are needed to get Rs = 1.5, then error might be very large. To determine the constraints of this method, errors have been reported according to changing resolution when quantitating proportionate as well as disproportionate overlapping chromatographic peaks (11,20). Peak area quantification was accurate within 1% error when Rs was >0.8 for two overlapping proportionate peaks (50:50 area ratio) (20). With overlapping peaks of area in proportion of 1:99, error was much higher at similar resolution (20). Depending on the case, some method development might be necessary to obtain a resolution around 0.8 before applying power transformation.
Direct Resolution Enhancement by Even Derivative Peak Sharpening
Using even derivatives to enhance chromatographic resolution is another example of directly increasing the resolution of chromatographic peaks post-data acquisition. The fundamental property of sharpening peaks is that for a symmetric peak function, the area under a derivative is zero (13). Real chromatographic peaks are rarely symmetric, but the area under a derivative for a tailing or fronting peak is negligible (on the order of 10-11 units, that is, signal • time). Therefore, if we add or subtract even time-derivatives of peaks from the raw chromatographic data, the peak areas should not change. The result is a sharper peak, which increases the chromatographic resolution between adjacent peaks. It is important to smooth the data so the noise is minimal before subtraction or addition. The idea can be expressed mathematically, as shown in equation 4:

where K2 and K4 are constant multipliers with consistent units to make the derivatives dimensionless. The user can empirically tune these values until the desired peak widths are obtained. Small dips are commonly observed at the front and back of the chromatographic envelope, but do not change the peak area or interfere with integration if properly included in integration (13). An Excel template was created to automate this process, such that a chromatogram could be exported and then resolved (13).
To visualize this technique, a simulated Gaussian peak with an area of 1 is shown in Figure 4a. The result of subtracting the second derivative and adding the fourth derivative (each with an appropriate multiplier) is shown in Figure 4b. The sixth derivative was also added, but its effect is negligible. The peak width is reduced and the peak height increased while the area remains = 1. Thus, the even derivative method is a peak-shaping protocol to make the peaks narrow. This method can operate on all components of a chromatogram simultaneously, unlike the modified power law where each peak has to be treated individually (20). In Figure 4c, a twin-column recycling HPLC chromatogram separating d3- and d6-benzenes from ordinary benzene is shown (13). In recycling HPLC, the analytes are continuously injected and detected; that is, they are recycled in the chromatograph until the desired resolution is obtained. For this separation, it takes about 1.5 h to separate deuterated benzenes completely (Figure 4d). Instead of waiting 1.5 h for baseline resolution, a faster approach would be to determine each peak area by equation 3. Figure 4c shows the peak sharpening of the fourth recycled chromatogram (segment IV from Figure 4d). From this point onwards, accurate peak area estimation (<~1% error) can be obtained even before the physical separation is complete. Error for peak area determination of two overlapping proportionate peaks was determined to be within 1% if the chromatographic resolution was >0.7 (13).
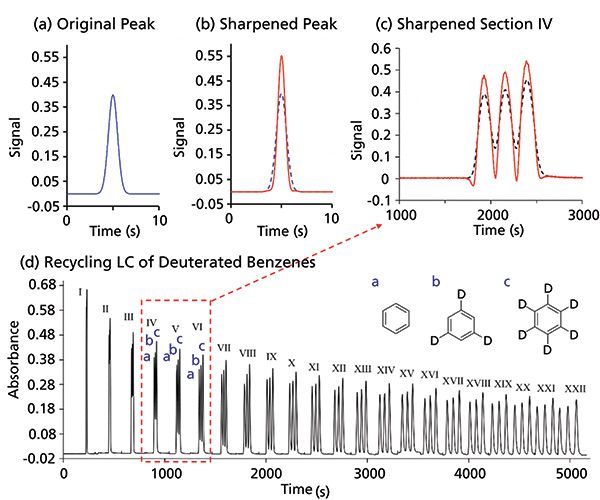
Figure 4: Sharpening peaks with even derivatives. (a) shows a simulated Gaussian peak (in blue). (b) shows the effect of sharpening the simulated peak (in blue) by reducing the peak width (in red). This is done by subtracting the second and adding the fourth derivatives with their appropriate multipliers. The area of the peak is conserved. (c) and (d) show the separation of an isotope mixture containing (a) benzene, (b) 1,3,5-benzene-d3, and (c) benzene-d6. See reference 13 for chromatographic information. The separation takes up to 1.5 h to get baseline resolution needed for quantitation. However, using even derivative peak sharpening (c), section IV (in black) can be baseline resolved (in red) increasing throughput by ~1 h. Adapted with permission from reference 13.
A Quick Comparison of Peak Resolution Methods
Figure 5 provides a quick overview of the four methods discussed above when multidimensional data are not available, or when not applicable. These techniques can be applied on any single-channel data in any mode of chromatography (GC, LC, or SFC), and in capillary electrophoresis with any detector. The original data (Figure 5) consist of six overlapping peaks with noise. The instrumental band broadening can be removed by FT deconvolution. As is evident, Figure 5a increases the resolution by removing the tailing caused by the instrument itself. The iterative curve fitting procedure can resolve the six peaks baseline with accurate areas as exponentially modified peaks (Figure 5b). MCR-ALS provides similar results to iterative curve fitting. However, it requires multidimensional data and does not need a peak model. In order to easily visualize all the six peaks, one can apply a positive integer power by raising the signal to power 3 (12) on Figure 5a. Finally, the first and second derivative sharpening method (13) can be applied on Figure 5a to make the peaks baseline for convenient integration. Further studies are underway to improve these resolution enhancing procedures.
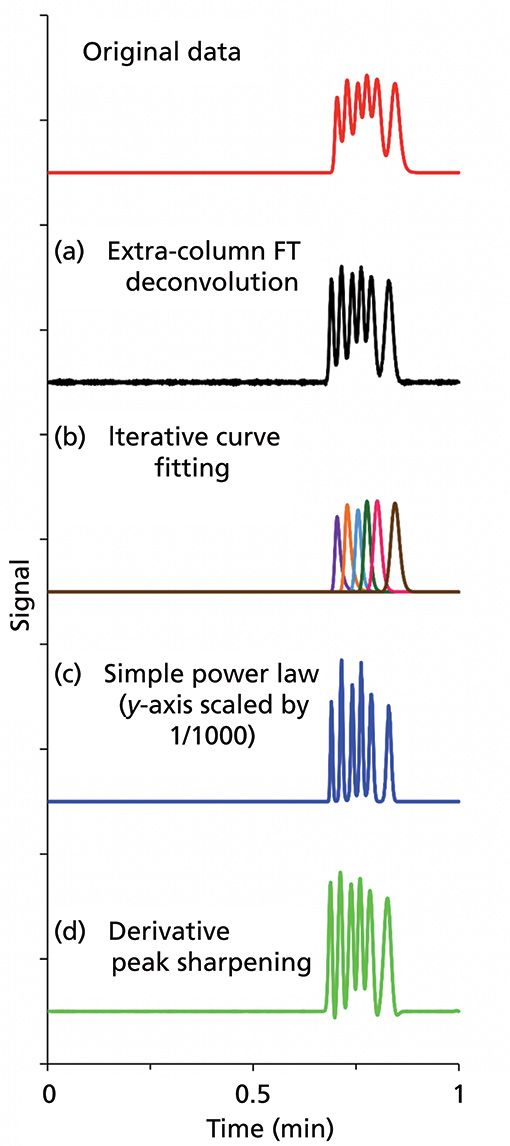
Figure 5: Overview of each signal processing technique. Original data simulated of six components partially separated in a under a minute. (a) Fourier-Transform Deconvolution: Dead volume of an Agilent 1200 HPLC was determined at 3 mL/min and used to remove the extracolumn band broadening. (b) Iterative Curve Fitting: The chromatogram was fitted using a bidirectional exponentially modified Gaussian model providing the extracted areas of each peak under the curve. (c) Simple power law: The data were raised to power = 3, then scaled down to fit in the same signal window as other methods. The modified power law could be used to quantitate the individual peak areas one at a time. (d) Derivative peak sharpening: Adding the first and subtracting the second derivatives with constants K1 and K2 of 0.0051 and 0.000005, respectively.
Conclusions
Resolution enhancement strategies seem to be the next step in improving chromatographic separations, not only to determine peak areas of overlapping peaks, but also to deconvolute system effects, reduce noise, and fix asymmetry. These strategies aim to increase throughput and offer cost-effective solutions compared to traditional method development. Their automation will surely make them extremely useful to the chromatography community, and hence this intelligent peak processing is the future of chromatography. In general, the techniques described in this review either remove extracolumn band broadening (Fourier transform deconvolution), extract peak area from under a curve (iterative curve fitting and multivariate curve resolution), or directly enhance chromatographic resolution (modified power law and even derivative peak sharpening). There are benefits and limitations to each technique; one might be more favorable than another for a specific application, and the users have to apply their own judgement on the choice of resolution enhancing methods.
Acknowledgements
The authors thank Yoachim Vanderheyden and Ken Broeckhoven for providing MATLAB figures for FT deconvolution (Figure 1). We also thank Prof. Thomas O'Haver for collaboration.
References
(1) S. Bruns, E.G. Franklin, J.P. Grinias, J.M. Godinho, J.W. Jorgenson, and U. Tallarek, J. Chromatogr. A 1318, 189–197 (2013).
(2) A.E. Reising, S. Schlabach, V. Baranau, D. Stoeckel, and U. Tallarek, J. Chromatogr. A 1513, 172–182 (2017).
(3) M.F. Wahab, D.C. Patel, R.M. Wimalasinghe, and D.W. Armstrong, Anal. Chem. 89, 8177–8191 (2017).
(4) F. Gritti and M.F. Wahab, LCGC Europe 31, 90–101 (2018).
(5) J.M. Davis and J.C. Giddings, Anal.Chem. 55, 418–424 (1983).
(6) Y. Vanderheyden, K. Broeckhoven, and G. Desmet, J. Chromatogr. A 1465, 126–142 (2016).
(7) N.A. Wright, D.C. Villalanti, and M.F. Burke, Anal. Chem. 54, 1735–1738 (1982).
(8) H. Parastar and R. Tauler, Anal.Chem. 86, 286–297 (2014).
(9) S.N. Chesler and S.P. Cram, Anal.Chem. 45, 1354–1359 (1973).
(10) A. De Juan and R. Tauler, Crit. Rev.Anal. Chem. 36, 163–176 (2006).
(11) M.F. Wahab, F. Gritti, T.C. O'Haver, G. Hellinghausen, and D.W. Armstrong, Chromatographia 82, 211–220 (2019).
(12) P.K. Dasgupta, Y. Chen, C.A. Serrano, G. Guiochon, H. Liu, J.N. Fairchild, and R.A. Shalliker, Anal. Chem. 82, 10143–10150 (2010).
(13) M.F. Wahab, T.C. O'Haver, F. Gritti, G. Hellinghausen, and D.W. Armstrong, Talanta 192, 492–499 (2019).
(14) D.C. Patel, M.F. Wahab, T.C. O'Haver, and D.W. Armstrong, Anal. Chem. 90, 3349–3356 (2018).
(15) S. Misra, M.F. Wahab, D.C. Patel, and D.W. Armstrong, J. Sep. Sci. 42, 1644–1657 (2019).
(16) J. Schenk, J.X. Mao, J. Smuts, P. Walsh, P. Kroll, and K.A. Schug, Anal. Chim.Acta 945, 1–8 (2016).
(17) M. Maeder, Anal. Chem. 59, 527–530 (1987).
(18) D.W. Cook, S.C. Rutan, C. Venkatramani, and D.R. Stoll, LCGC North Am. 36, 248–255 (2018).
(19) https://www.shimadzu.com/an/literature/hplc/jpl217011.html (Accessed 24 April 2019).
(20) G. Hellinghausen, M.F. Wahab, and D.W. Armstrong, J. Chromatogr. A 1574, 1–8 (2018).
M. Farooq Wahab is a Research Engineering Scientist at the University of Texas, in Arlington. Garrett Hellinghausen is a PhD student at the University of Texas, in Arlington. Daniel W. Armstrong is the Welch Distinguished Professor of Chemistry at the University of Texas, in Arlington. Direct correspondence to Daniel Armstrong at sec4dwa@uta.edu

Understanding FDA Recommendations for N-Nitrosamine Impurity Levels
April 17th 2025We spoke with Josh Hoerner, general manager of Purisys, which specializes in a small volume custom synthesis and specialized controlled substance manufacturing, to gain his perspective on FDA’s recommendations for acceptable intake limits for N-nitrosamine impurities.









