Micro-Pillar Array Column Separations for Proteomics of Liver Organoids
Organoids are laboratory-grown three-dimensional (3D) models of organs and are emerging tools for studies into developmental biology, drug discovery, and toxicology. Organoids are complex biological materials, and proteomics studies of organoids can benefit from applying high-resolution chromatography devices before mass spectrometric analysis. Micro-pillar array columns have been shown to provide excellent resolution of peptide products of proteolytic digestion of proteins for bottom-up proteomics.This article describes a workflow incorporating a micro-pillar array column for mapping the proteome of human stem cell-derived liver organoids (sample preparation using a SPEED protocol) using trapped ion mobility time-of-flight mass spectrometry (timsTOF-MS).
Organoids are three-dimensional laboratory-grown cell structures, developed from stem cells into models of, for example, liver, pancreas, kidney, and brain (1). Organoids can arguably serve as more relevant models compared to animal models, which often do not satisfactorily represent human physiology in, for example, the preclinical stages of drug development (2). As organoids can be grown from the cells of an individual patient, or with introduced patient specific mutations, they can serve as important tools for personalized medicine.
3d rendered illustration of the male liver | Image Credit: © SciePro - stock.adobe.com
Organoids are complex biomaterials, often consisting of several cell types, mimicking aspects of the composition and morphology of the model organ. Organoids are still in the early stages of their development but are advancing in complexity and utility.
A key necessity for organoid engineering is the ability to track their composition and traits. Taking liver organoids as an example, their ability to metabolize drugs can serve as an indicator of their maturity and relevance for testing novel therapeutics. Other frequently applied analytical tools include immunofluorescence and analyzing their transcriptome, genome, metabolome/lipidome, and proteome. Separation science and mass spectrometry (MS) are key tools for some of this work (3).
This article describes a proteomic study of liver organoids, generated from human-induced pluripotent stem cells, with the post-differentiation maturation steps aiming to increase metabolic activity of liver organoids developed in-house (4).
This study used micro-pillar array columns that we have previously applied for single-dimension separations of biomedical samples (5). The pillar arrays of the micro-pillar array columns used are highly ordered, providing separations at relatively low pressures, which allows long columns for high-resolution analysis to be used. The micro-pillar array column was coupled with high-resolution mass spectrometry (HRMS), for separation and mass analysis following sample preparation using SPEED digestion (6).
Materials and Methods
Small Equipment and Consumables
Falcon centrifuge tubes (15 mL) were from VWR. Safe lock tubes (1.5 mL) were purchased from Eppendorf, while the low-binding safe lock tubes (1.5 mL) were purchased from Sarstedt. Omix C18 10 μL pipette tips were purchased from Agilent.
A PSC24 thermo-shaker was used for all the incubation steps and was purchased from Grant Instruments. The test tube shaker was from Heidolph instruments. A Mini Star minicentrifuge was from VWR. Vacuum centrifuges (concentrator plus and centrifuge 5424 R) were purchased from Eppendorf.
Reagents and Solutions
High performance liquid chromatography (HPLC) water, acetic acid (100%), acetonitrile, and formic acid (FA) (≥ 99%) were purchased from VWR. Trifluoroacetic acid (TFA) (99.8%), iodoacetamide (IAM), DL-dithiothreitol (DTT), and Trizma base (Tris base) were from Sigma-Aldrich. Pierce HeLa protein digest standard and Gibco Dubecco’s phosphate-buffered saline (DPBS) were purchased from Thermo Fisher Scientific. Trypsin Gold, mass spectrometry-grade, was purchased from Promega. A solution of 2 M Trisbase was prepared by dissolving 2.4 g in 10 mL of water. The trypsin stock solution of 1 μg/μL was prepared by dissolving 100 μg of trypsin in 100 μL of 50 mM acetic acid. By dissolving 77.2 mg DTT in 1000 μL water and 92.5 mg IAM in 500 μL water, 0.5 M DTT and 1 M IAM were prepared, respectively. For the micro solid-phase extraction (mSPE), the conditioning and elution solutions were 0.1% TFA in 50% acetonitrile, and the equilibration and washing solution were 0.1% TFA in water.
Samples
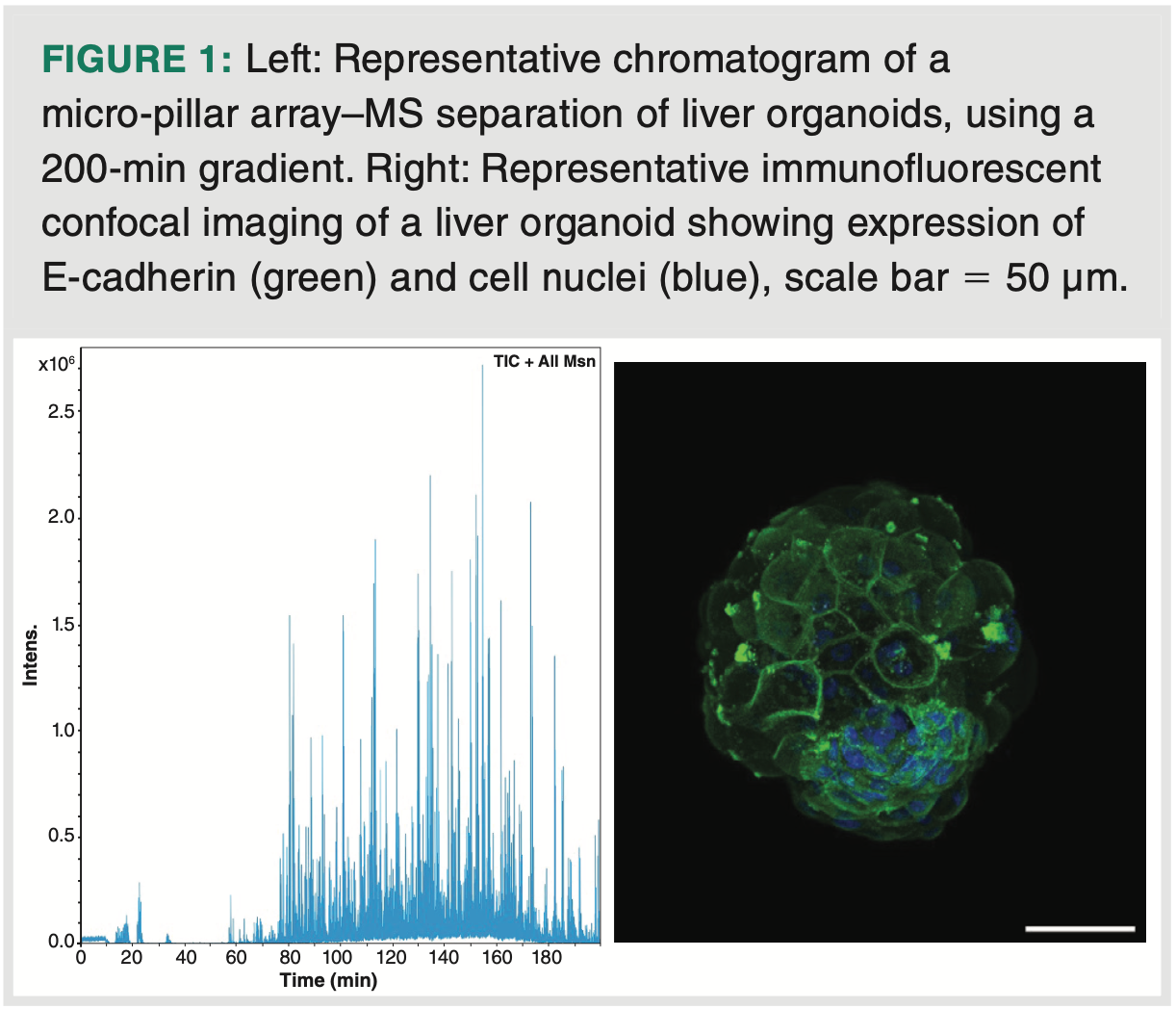
The analytical samples consisted of liver organoids generated from iPSC cell line (WTSIi013-A, Wellcome Trust Sanger Institute) by three differentiation protocols. Protocol 1 represents a standard differentiation protocol (7); protocol 2 and 3 represent differentiation protocols that lead to more metabolic mature liver organoids (4). Each sample consisted of 20–30 organoids (see Figure 1 for image of a liver organoid). Each of the organoid samples was thawed on ice for approximately 1 h and washed with 200 μL ice-cold DPBS and suspended by gently rotating the vial. The samples were centrifuged at 400 × g /4 °C /5 min, before discarding the supernatant.
Sample Preparation
The samples were prepared following the protocol for sample preparation of easy extraction and digestion (SPEED) by Doellinger et al. (6). The organoids were lysed with neat TFA, in a 1:4 ratio (sample:TFA). To neutralize the sample, 2M Tris base was added in a volume ten times the volume of TFA. Tris(2-carboxyethyl)phosphine (TCEP) and 2-choloroacetamide (CAA) (reduction and alkylation reagents in the original protocol) were replaced with DTT and IAM. DTT was added to a final concentration of approximately 10 mM, and incubated at 700 rpm at 56 °C for 25 min. Subsequently, IAM was added to a final concentration of 20 mM and incubated at 700 rpm at room temperature for 30 min in the dark. To quench the alkylation, 5 mM DTT was added and incubated at 700 rpm at room temperature for 15 min. The samples were diluted 1:5 with H2O. Digestion was promoted by adding 0.5 μg of trypsin, with an overnight incubation at 700 rpm at 37 °C. TFA was added to a final concentration of 2%.
The samples were desalted using an Omix Tip with C18 material embedded in the tip. The Omix Tip was conditioned by aspirating 30 μL (3 × 10 μL) conditioning solution, before discarding the solution. The same procedure was repeated for the equilibration using an equilibration solution (0.1% TFA in water). The sample was loaded by pipetting 10 μL at a time and slowly eluting the sample. To rinse the Omix Tip, 30 μL (3 × 10 μL) washing solution (0.1% TFA in water) was aspirated and discarded. The sample was eluted with 10 μL elution solution (0.1% TFA in 50% acetonitrile) in a new, clean vial. The samples were dried at room temperature for 60 min and reconstituted in 10 μL 0.1% FA.
LC–MS Instrumentation
The samples were analyzed by nanoflow-LC–MS using a timsTOF Pro (Bruker Daltonik), which was coupled online to a nanoElute nanoflow liquid chromatography system (Bruker Daltonik) via a CaptiveSpray nanoelectrospray ion source (Bruker Daltonik). The peptides were separated on a 200 cm μPAC column (PharmaFluidics, now acquired by Thermo Fisher). Mobile phase A contained water with 0.1% formic acid, and acetonitrile with 0.1% formic acid was used as mobile phase B. The peptides were separated by a gradient from 2–35% mobile phase B over 200 min at a flow rate of 300 nL/minute. MS acquisition was performed in DDA-PASEF mode. The capillary voltage was set to 1.5 kV, with a mass range of 100 to 1700 m/z. The number of PASEF ranges was set to 20, with a total cycle time of 1.16 s, charge up to 5, target intensity of 20,000, intensity threshold of 1750, and active exclusion with release after 0.4 min. An inversed reduced trapped ion mobility (TIMS) (1/k0) of 0.85–1.40 Vs/cm2 was used, with a range time of 100 ms, an accumulation time of 100 ms, a duty cycle of 100%, and a ramp rate of 9.51 Hz. Precursors for data-dependent acquisition were fragmented with an ion mobility-dependent collision energy, which was linearly increased from 20 to 59 eV.
Database Search
The LC–MS data were searched against the human Uniprot database (20,431 entries), with PEAKS X+ software version 10.5 (Bioinformatics Solutions). The following parameters were used: digestion enzyme, trypsin; maximum missed cleavage, 2; fragment ion mass error tolerance, 0.03 Da; parent ion error tolerance, 15 ppm. Oxidation of methionine and acetylation of the N-terminus were specified as variable modifications, and carbamidomethylation of cysteines as fixed modification. The maximum number of post-translational modifications (PTMs) per peptide was set to three. A false-discovery rate (FDR) of 1% was applied to the datasets.
Label-Free Quantitation
For label-free quantification (LFQ) using PEAKS X+, four biological replicates of protocol 2, protocol 3, and a control were used. The following parameters were applied on peptide features: quality ≥ 5, charge: 2–5, and on protein: significance: 5% FDR, fold change ≥ 2, significance method ANOVA with at least two used peptides. Twenty-five internal standard proteins were used for normalization.
Results and Discussion
Micro-Pillar Array–MS Workflow
The benefits of SPEED include fast detergent-free lysis, minimal sample handling, and an easy-to-follow protocol. Following SPEED preparation of the limited organoid samples (Figure 1), the prepared samples were subject to HRMS using timsTOF–MS, downstream to LC separation using a 200 cm micro-pillar array column and a solvent gradient over 200 min (Figure 1). Even with this long column format, the micro-pillar array column used had a back pressure of only 140 bars. Some practical challenges were that the nanoflow-LC–MS setup of the core facility featured connection fittings that were not directly compatible with this micro-pillar array column, requiring two transition connectors, which can potentially contribute to dead volumes. With the methodology, approximately 3000 different proteins were identified and measured on each organoid sample comprising 20–30 organoids.
Analysis of Liver Organoids
Liver organoids are developed for the in vitro modelling of liver disease, drug development, and testing studies, as well as basic research of liver physiology. By analyzing in-house developed organoids, we hypothesized that liver organoids generated by protocol 2 would have increased metabolic activity, particularly cholesterol biosynthesis, increased tricarboxylic acid (TCA) cycle and oxidative pathway, fatty acids beta-oxidation, and gluconeogenesis, when compared to the standard protocol; while the cells from protocol 3 would have increased glucose uptake and glycogen storage, lipogenesis, and bile acid synthesis.
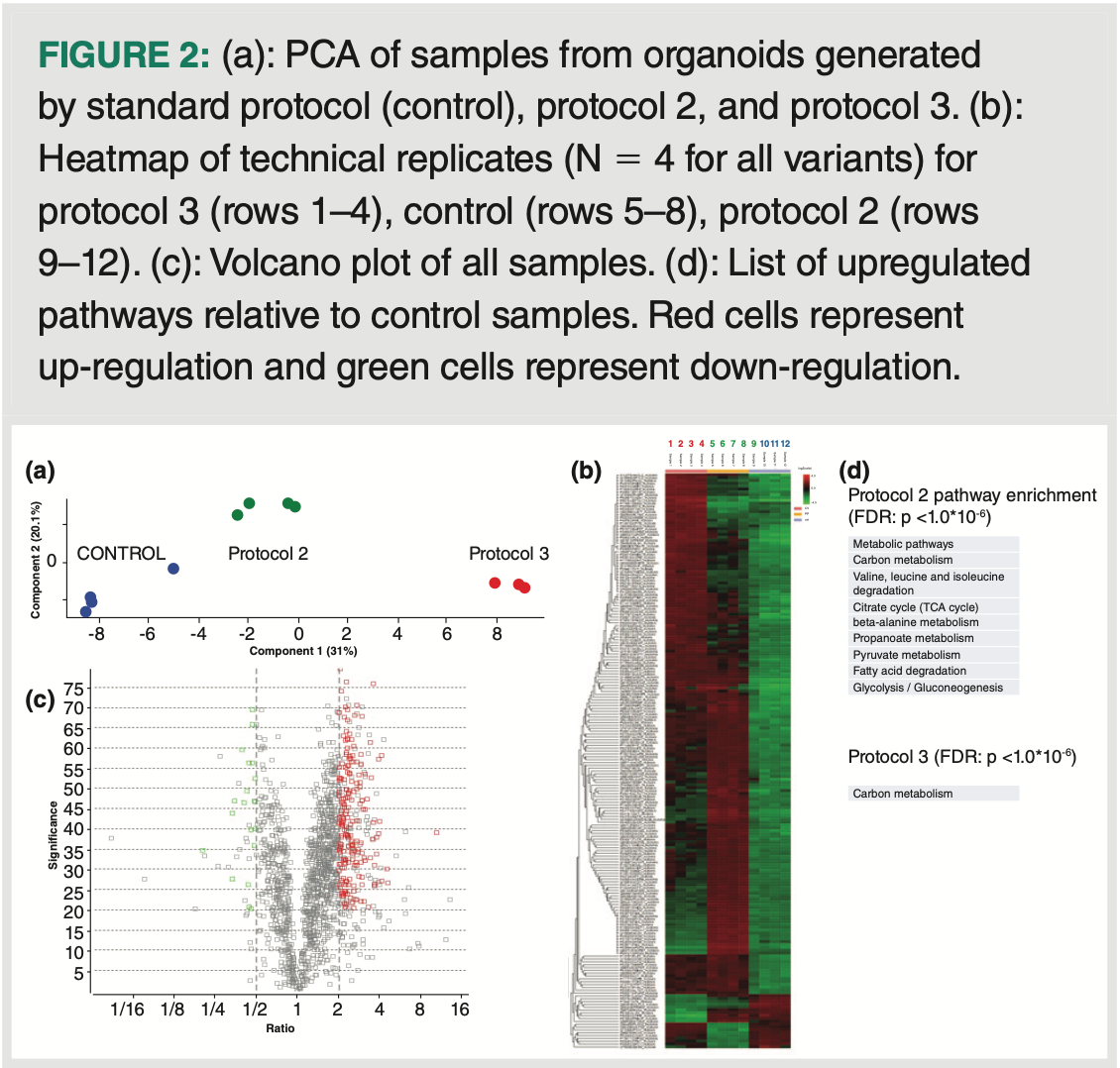
Principal component analysis (PCA) of data generated using the method described (N = 4 replicates per group) demonstrated clear differences between the three groups (Figure 2[a]). A heatmap of the samples further visualizes the differences among the three groups as
well as the relative homogeneity within the technical replicates, that is, the colour uniformity between the different columns within each group category (Figure 2[b]). Specific species that were up/downregulated were visualized with a volcano plot, that is, plotting statistical significance vs. fold-change (Figure 2[c]). Species that were significantly regulated (FDR = 5%, fold change 2) were subsequently analyzed regarding the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment; as presented in Figure 2(d), samples from protocol 2 largely met the predicted traits, for example, upregulation related to TCA cycle, fatty acid metabolism, and gluconeogenesis. Less information was obtained for the protocol 3 samples. Nonetheless, despite the small sample size, the methodology allowed a clear distinction between the three different liver organoid groups.
Conclusions
The analytical workflow described allows analysts to unequivocally group liver organoids developed by three differentiation protocols. This analytical pipeline provides a comprehensive characterization of metabolism and phenotype of liver organoids, reflecting their different maturation status. Results were generated from the limited number of organoids (20–30 organoids per replicate)—similar to routine but less informative assays such as immunofluorescent microscopy or quantitative polymerase chain reaction (qPCR). It can therefore be broadly included in the analytical toolbox for characterization of stem cell-derived organoids.
Acknowledgements
This work was supported by the Research Council of Norway through its Centre of Excellence scheme, project number 262613, and project number: 295910 (National Network of Advanced Proteomics Infrastructure). Support was also provided by UiO:Life Science and the Olav Thon Foundation. We are grateful for supporting data analysis by Dr. Tuula Nyman and technical assistance by Dr. Manuel Ramirez Garrastacho.
Author Statement
AA and LM share first authorship.
References
(1) Zhao, Z.; Chen, X.; Dowbaj, A. M.; et al. Organoids. Nat. Rev. Methods Primers 2022, 2, 94. DOI: 10.1038/s43586-022-00174-y
(2) Li, M.; Izpisua Belmonte, J. C. Organoids-Preclinical Models of Human Disease. N. Engl. J. Med. 2019, 380, 569–579. DOI: 10.1056/NEJMra1806175
(3) Kogler, S.; Kømurcu, K. S.; Olsen, C.; et al. Organoids, Organ-on-a-Chip, Separation Science and Mass Spectrometry: An Update. Trends in Anal. Chem. 2023, 161, 116996. DOI: 10.1016/j.trac.2023.116996
(4) Aizenshtadt, A.; Liu, S.; Wang, C.; et al. In Vitro Zonation of Human iPSC-Derived 3D Liver Organoids. Hepatology 2022, 76, S1–S1564. Abstract no. 4116.
(5) Berg, H. E.; Halldórsson, S.; Bakketeig, E. A.; et al. Micro-Pillar Array Columns (μPAC): An Efficient Tool for Comparing Tissue and Cultured Cells of Glioblastoma. J. Chromatogr. Open 2022, 2, 100047. DOI: 10.1016/j.jcoa.2022.100047
(6) Doellinger, J.; Schneider, A.; Hoeller, M.; Lasch, P. Sample Preparation by Easy Extraction and Digestion (SPEED) - A Universal, Rapid, and Detergent-free Protocol for Proteomics Based on Acid Extraction. Mol. Cell. Proteomics 2022, 19, 209–222. DOI: 10.1074/mcp.TIR119.001616
(7) LaLone, V.; Aizenshtadt, A.; Goertz, J.; et al. 2023. Quantitative Chemometric Phenotyping of Three-Dimensional Liver Organoids by Raman Spectral Imaging. Cell Rep. Methods. 2020, 3, 100440. DOI: 10.1016/j.crmeth.2023.100440
About the Authors
Aleksandra Aizenshtadt is a postdoctoral research fellow at the Hybrid Technology Hub, University of Oslo.
Lise Midtøy is a master’s student in the Bioanalytical Chemistry group at the Department of Chemistry, University of Oslo.
Bernd Thiede is head of the Section for Biochemistry and Molecular Biology, Department of Biosciences, University of Oslo.
Stefan Krauss is a professor at the Institute of Basic Medical Sciences, University of Oslo, and the director of the Hybrid Technology Hub.
Hanne Røberg-Larsen is an associate professor and head of the Bioanalytical Chemistry group at the Department of Chemistry, University of Oslo.
Steven Ray Wilson is a professor at the Department of Chemistry, University of Oslo, and a PI in the Hybrid Technology Hub.

New Method Explored for the Detection of CECs in Crops Irrigated with Contaminated Water
April 30th 2025This new study presents a validated QuEChERS–LC-MS/MS method for detecting eight persistent, mobile, and toxic substances in escarole, tomatoes, and tomato leaves irrigated with contaminated water.
Accelerating Monoclonal Antibody Quality Control: The Role of LC–MS in Upstream Bioprocessing
This study highlights the promising potential of LC–MS as a powerful tool for mAb quality control within the context of upstream processing.
University of Tasmania Researchers Explore Haloacetic Acid Determiniation in Water with capLC–MS
April 29th 2025Haloacetic acid detection has become important when analyzing drinking and swimming pool water. University of Tasmania researchers have begun applying capillary liquid chromatography as a means of detecting these substances.
Prioritizing Non-Target Screening in LC–HRMS Environmental Sample Analysis
April 28th 2025When analyzing samples using liquid chromatography–high-resolution mass spectrometry, there are various ways the processes can be improved. Researchers created new methods for prioritizing these strategies.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)