Innovation in Nutrition and Food Analysis: The FoodOmicsGR_Research Infrastructure
There has been an increased interest in monitoring food quality and safety, and their important role in relation to human life, wellness, and health. Existing regulations dictate how certain technological processes should be performed to assess food safety and quality by monitoring food components and the potential presence of contaminants. Although the “classical” targeted analytical perspective is still the major route, holistic approaches are gaining more momentum in characterizing food through “omics” technologies because this can provide novel unexpected markers of food quality.
Foodomics (1) combines food and nutrition sciences with advanced analytical techniques and bioinformatics to characterize the food composition or food consumers’ biofluids, and also to address new challenges such as authentication, traceability, improvement of food produce, food quality, and nutritional value. This holistic approach employs a cutting‑edge research infrastructure, where big data are produced that generate new knowledge and highlight previously unknown associations of biomolecules with the studied phenotype.
The potential application of foodomics is very wide and includes the following areas: food characterization and differentiation; food traceability and authenticity; adulteration control; identification of origin (geographic or genetic); monitoring of maturation; assessing freshness; linking organoleptic characteristics with metabolite composition; supporting health claims of novel functional (super)foods; understanding mechanisms of toxicity; development of a personalized dietary intervention for patients with special needs, for example, food allergies; and identifying legitimate or non-legitimate interventions (hormone treatment, use of fertilizers in crops). Ultimately, this leads to biomarker discoveries that can reveal the beneficial effects of nutrition on human health. Such data offer key elements to support marketing and brand naming. Hence, products of high economic value, such as products in the category of protected designation of origin (PDO) and protected geographical indication (PGI), can be characterized extensively and can be more effectively protected. Overall, foodomics as an omics approach can provide new knowledge and reveal biochemical mechanisms that describe the advantageous or adverse effects of food components (2). Figure 1 provides an illustration of the fields and the various subfields and applications involved in foodomics.
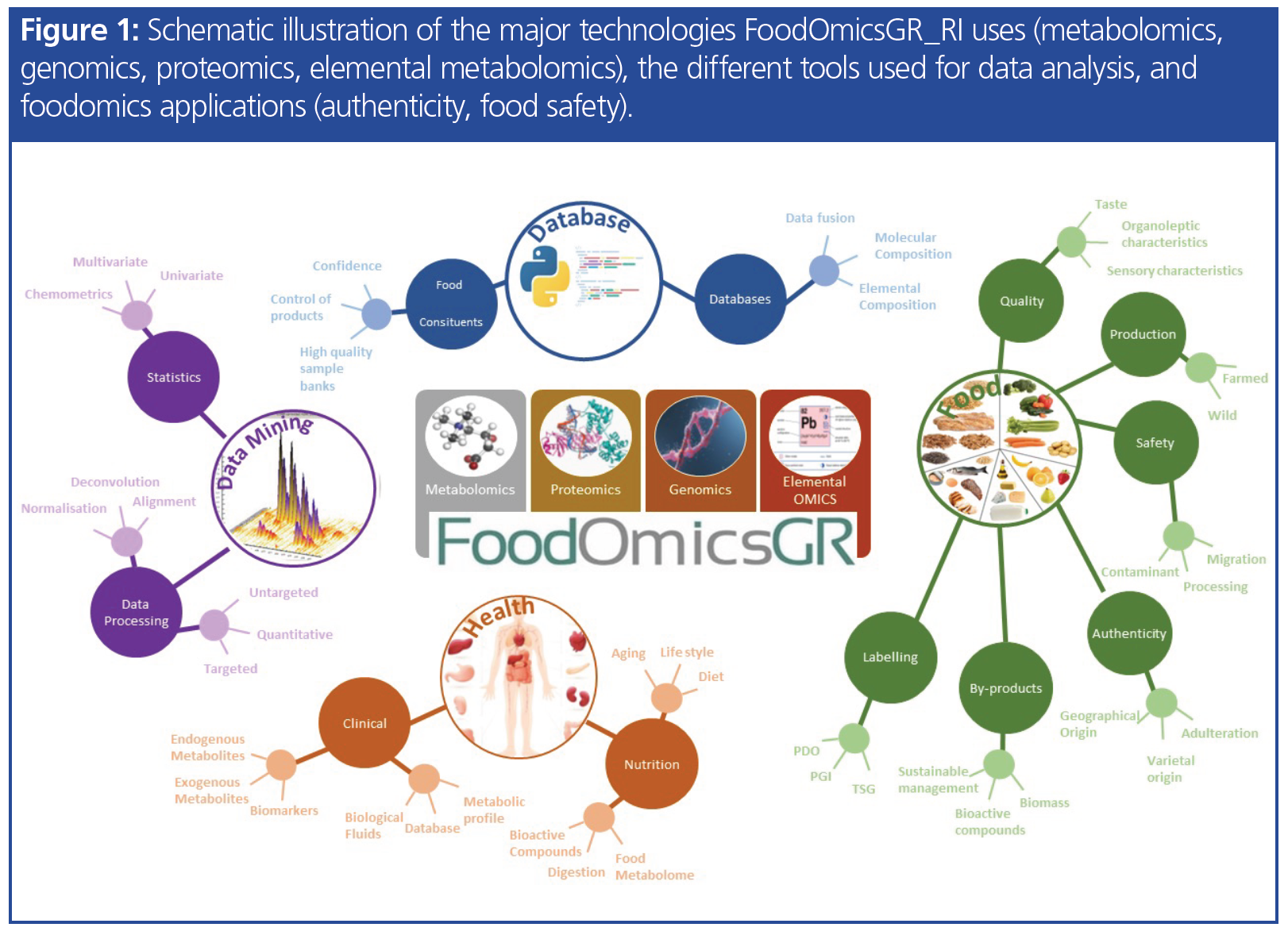
The Project: FoodOmicsGR_RI
FoodOmicsGR_RI is a national research infrastructure that aims to perform and aid omics research in the agri-food sector in the Greek research environment. This field is of high importance for the country and its agriculture because of the unique landscape Greece exhibits that results in a diverse and rich portfolio of local products and foods. Therefore, the central scope of FoodOmicsGR_RI is to support the Greek agri-food sector by generating robust data on the composition and nutritional value of the local produce, thereby increasing a product’s position, market demand, and provide a higher revenue for the producers.
The project comprises eight Greek universities and research centres. Analytical groups and food specialist groups from the universities of Athens, Crete, the Aegean, Ioannina, the Agricultural University of Athens, the International Hellenic University, and the Biomedical Research Center of the Academy of Athens comprise a team of 60 staff scientists and 40 newly recruited researchers from 20 scientific disciplines. BIOMIC_AUTH, a new interdisciplinary laboratory of the Aristotle University Thessaloniki (http://biomic.web.auth.gr/), with great expertise in bioanalytical chemistry, is coordinating the project. More details can be found on the website (http://foodomics.gr/).
Cutting-edge research technologies include high performance liquid chromatography (HPLC), mass spectrometry (MS), nuclear magnetic resonance (NMR) spectroscopy, and inductively coupled plasma–mass spectrometry (ICP-MS) instrumentation and software. These are deployed to cover a wide area of applications such as bioanalysis, metabolomics, genomics, proteomics, and bioinformatics. The capital instrumentation of FoodOmicsGR_RI laboratories includes:
- Five quadrupole time-of-flight (QTOF)-MS instruments (two trapped ion mobility mass spectrometry systems [TIMS]-TOF-MS)
- Three matrix-assisted laser desorption–ionization (MALDI)-TOF-MS instruments
- Four ultrahigh-pressure liquid chromatography (UHPLC)–orbital trap high resolution mass spectrometry (HRMS) instruments (one nano‑LC)
- More than 10 UHPLC triple quadrupole MS/MS systems
- Three HPLC–MS single quadrupole systems
- Supercritical fluid chromatography (SFC)–UV/MS system
- More than eight gas chromatography (GC)–MS systems (single quadrupole)
- Four GC–MS/MS systems (triple quadrupoles)
- NMR spectroscopy (600 MHz, 500 MHz, and 400 MHz NMR)
- Two ICP-MS, two ICP-optical emission spectrometry (OES) instruments, next generation sequencing (NGS), and several other instruments.
As well as the analytical sciences, the consortium has a diverse and in-depth practical knowledge in the food sciences, namely fishery, apiculture, animal husbandry, plant growth, vinology, environmental control, natural products, food chemistry and technology, dairy product biochemistry, microbiology, and human exercise biochemistry. Researchers have gained experience from the analysis of thousands of samples of various origin by either untargeted or targeted metabolomics using HRMS; this experience includes reporting solid quantitative data in compliance with validation guidelines. FoodOmicsGR teams collaborate with the industry and the private sector (food producers, contract research organization [CRO] laboratories, life science companies, and analytical instrument vendors). In addition to that, the groups liaise with international research organizations, regulatory bodies, and consortiums (Eurachem, NIST, mQACC, Norman network, JRC-IRMM, and other key players).
FoodOmicsGR_RI offers:
- Consultancy for the design of experiment (DoE) for the foodomics and nutritional studies
- Genomic, proteomic, and metabolomic analysis of foods
- Quantitative analysis of key metabolites (nutrients, contaminants) in foods
- Big data processing and advanced statistical analysis.
FoodOmicsGR_RI aims to support R&D studies on food metabolomics, proteomics, and genomics applications, offering a “one‑stop‑shop” model to stakeholders and users.
The consortium partners offer expertise across major research lines related to the needs of the Greek agri-food domain. These can be grouped to the following research axes: 1) developing high-quality databases of Greek foods, 2) employing “omics” technologies to study local products and promote authenticity/traceability control, 3) highlighting unique traits of Greek products, 4) studying the effect of diet on health and well-being, 5) re-using and creating value from agri-food waste, and 6) studies on food safety assessment.
Key R&D axes of the consortium include: 1.Greek Food Composition Databases: One striking example of the portofolio is building in-house high-quality sample libraries establishing and enhancing the nutritional value of Greek foods. The group has published a number of topical reviews on food metabolomics such as olive oil (3), grapes, marc spirits, and wine (4). The group also generates compositional databases of the molecular composition of Greek agricultural products. The tools used for this purpose are data- and text-mining of the scientific literature (developing bespoke Python algorithms). The data are soon to be made publicly available on the project webpage.
2. Food Analysis, Traceability, and Control of Geographical Origin: FoodOmicsGR_RI research teams have developed and practiced MS and NMR methods for a variety of foods. Untargeted LC–HRMS methods combined with chemometric approaches complement traditional targeted LC–MS analysis (5,6) to study food authenticity.
The consortium has developed MS‑based workflows for food traceability and geographical origin, such as for olive oil, wine, dairy products, and honey. For example, classification of apple varieties was achieved based on volatiles analysis by headspace–solid‑phase microextraction (HS–SPME)–GC–MS (7), while the geographical origin of grapes was assessed using a hydrophilic interaction liquid chromatography (HILIC)–MS/MS method (4).
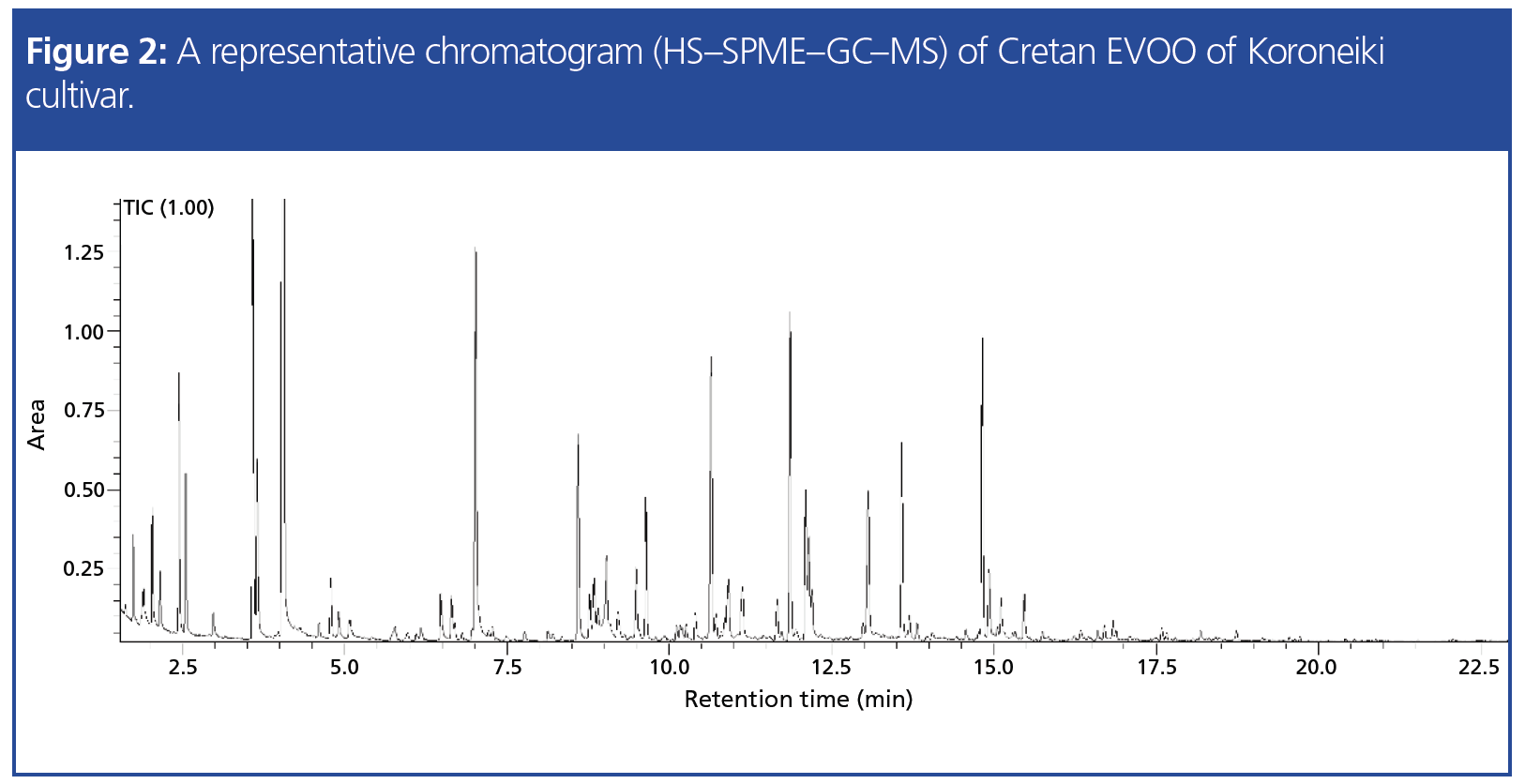
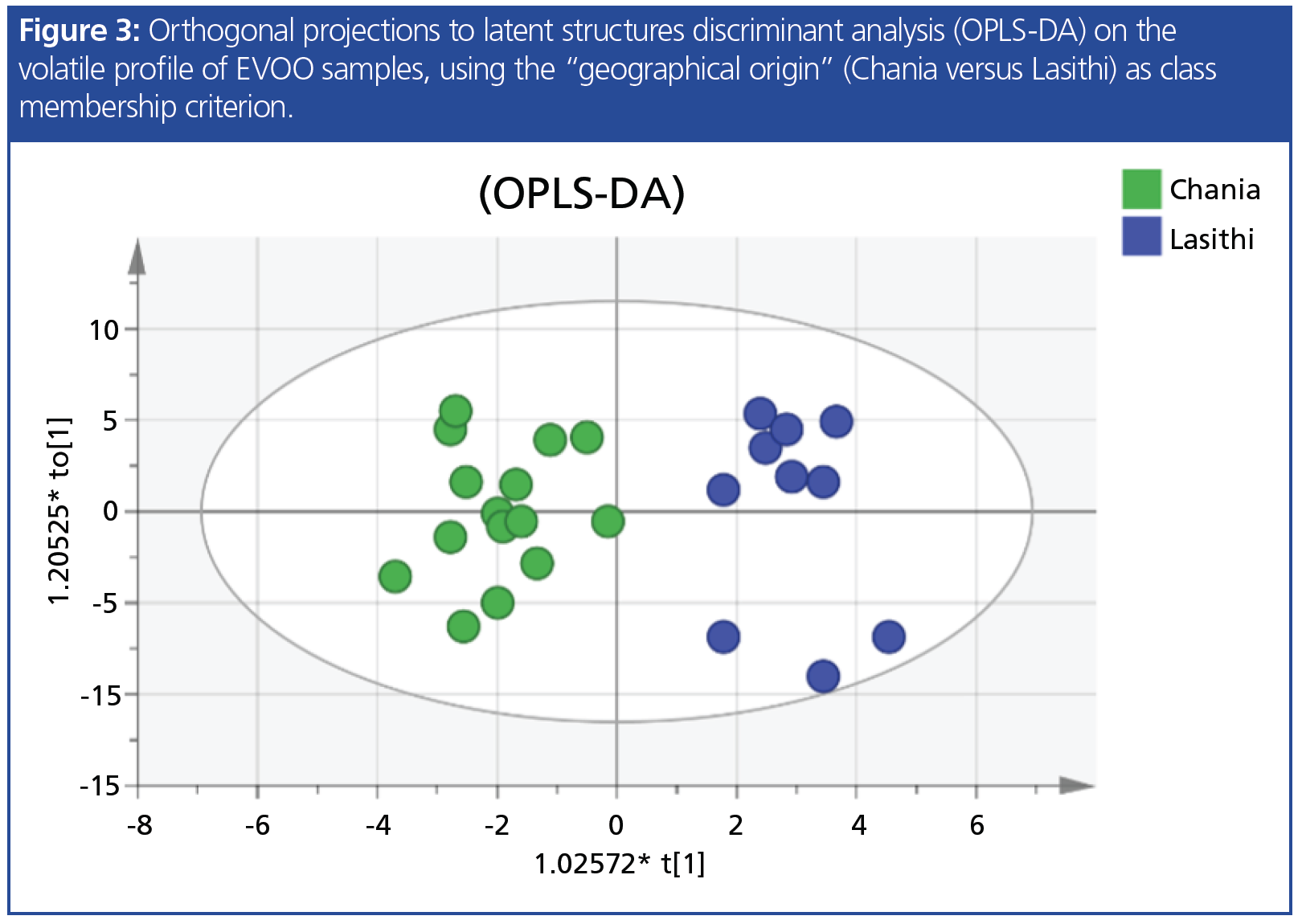
In another study conducted in the laboratory, 63 Cretan virgin olive oils were discriminated according to their geographical origin based on HS–SPME–GC–MS volatile organic compound (VOC) analysis. A representative gas chromatogram of VOCs from the Koroneiki cultivar is shown in Figure 2. The impact of geographical origin on volatile profile is illustrated in a orthogonal projections to latent structures discriminant analysis (OPLS-DA) model (Figure 3) where samples of different origin are visually separated.
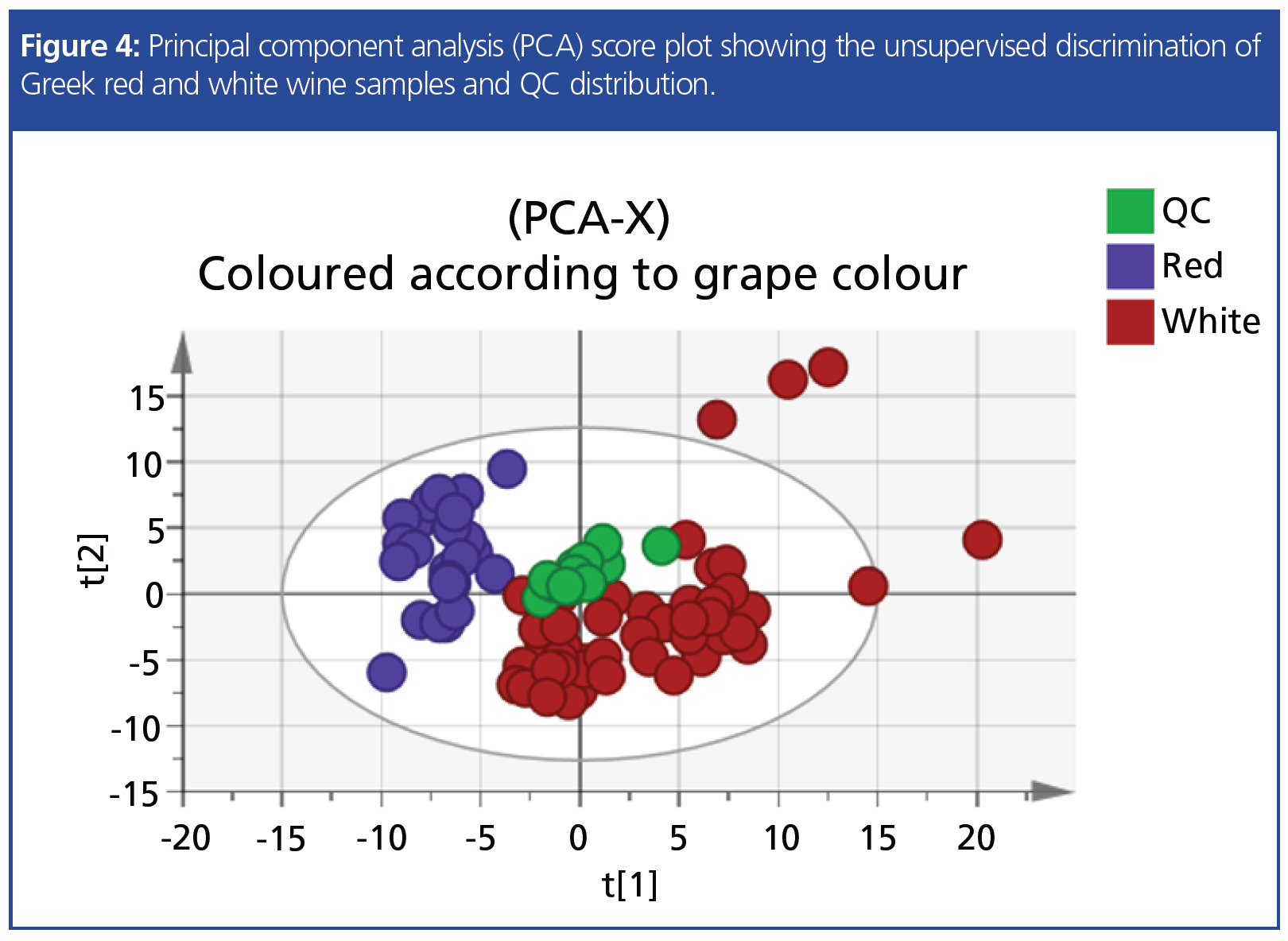
Another interesting example is an untargeted HS–SPME–GC–MS method that was applied for the classification of Greek wines based on their volatile composition. DoE assisted in the optimization of the analysis, while the sample collection consisted of 74 samples of nine different Greek grape varieties and six different regions. Preliminary results indicated that the classification of samples according to grape colour and variety is achievable (Figure 4).
3. Quality Control (QC) and Targeted MS Methods: FoodOmicsGR_RI laboratories have taken key steps and have extensive practical experience in the development of QC protocols for LC–MS-based metabolomics (8,9) for the validation of research findings. The group has studied the effect that sample preparation methods have on the obtained “metabolome” of different samples (10).
After two decades of metabolomics, there is a growing shift towards targeted quantitative methods, as recently reviewed by the group (11). The consortium has developed and validated targeted methods using GC–MS/MS or LC–MS/MS, providing robust and reproducible results for a set of metabolites; these include amino acids (12), organic acids, bile acids, or a wider group of up to 120 metabolites from different small molecule classes (13). Representative examples include studies of royal jelly (13,14), wine (15), and carobs, in addition to biological samples from nutritional studies.
4. Studies on Nutrition and the Nutritional Value of Foods: As stated previously, FoodOmicsGR_RI develops new tools to highlight the nutritional value of Greek foods; this can be attained by performing intervention studies where Greek traditional foods are used in the prevention of chronic diseases and to promote health claims. More specifically, the group monitors the effect of food ingredients at the genomic/transcriptomic/proteomic/metabolic/elemental level, followed by validation of initial findings. Ultimately the group develops proof-of-concept studies on food bioavailability on human organs and tissues. For example, the effect of a carob diet in the metabolome of biological samples of animal models has been studied (16). The facility has expertise in profiling biological fluids, such as urine, plasma, faeces, saliva, and tissues (liver, kidney, brain). Characteristic studies include the efffect of diet on human and animal models (16,17,18,19).
Other examples of nutritional studies include collaborations with partners from the private sector to improve health claims of novel superfoods from the Greek flora and marine organisms (that is, use of probiotics to maintain the balance of gut microbiota). Phenolic-rich water extracts from olives and their effect on animals and human health has also been studied. The group has investigated the role of novel animal feed enriched in chelated metals (Cu, Mn, Zn) and identified disease biomarkers in biofluids from sows after livestock intake (Project FITSOW) (20). A key goal is to offer the knowledge that could provide the base for the development of personalized diet based on observations of omics data integration. Such diets could improve health, well-being, and prevent the onset of disease.
5. Assessment of Food Safety
Food safety from chemical contaminants in foodstuffs as a result of environmental contamination, packaging, and storage processes is a major concern in the food market. To this end, the group has studied the migration of an endocrine disruptor (bisphenol A, [BPA]), from baby bottles into the aqueous content of foods or canned food (21). Also, non-intentionally added substances (NIAS) incorporated into the food chain have been studied by various analytical methods, for example, monitoring phthalates (22), oligomers, and other NIAS (23).
Conclusions
FoodOmicsGR_RI aims to characterize unique Greek products and highlight their value and quality using cutting-edge omics technology. The facility promotes and reinforces the collaboration between consortium partners, but, more importantly, it aims to act as a liaison focal point and link the research with the private sector and the producer. Overall, the group’s activity and R&D efforts help to increase the visibility and the market value for the investigated products, support brand naming, and generate higher revenue for the producers and the national economy.
Acknowledgement
We acknowledge the project “FoodOmicsGR_RI Comprehensive Characterization of Foods” (MIS 5029057), which is implemented under the action “Reinforcement of the Research and Innovation Infrastructure”, funded by the Operational Programme Competitiveness, Entrepreneurship, and Innovation (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund).
References
- V. García-Cañas, C. Simó, M. Herrero, E. Ibáñez, and A. Cifuentes, Anal. Chem. 84, 10150 (2012).
- A. Valdés, A. Cifuentes, and C. León, TrAC Trends in Analytical Chemistry 96, 2 (2017).
- A. Lioupi, N. Nenadis, and G. Theodoridis, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1150, 122161 (2020).
- D. Diamantidou, A. Zotou, and G. Theodoridis, Metabolomics 14, 159 (2018).
- H. Gika, C. Virgiliou, G. Theodoridis, R.S. Plumb, and I.D. Wilson, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1117, 136 (2019).
- H.G. Gika, I.D. Wilson, and G.A. Theodoridis, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 966, 1 (2014).
- E. Aprea, H. Gika, S. Carlin, G. Theodoridis, U. Vrhovsek, and F. Mattivi, J. Chromatogr. A 1218, 4517 (2011).
- O. Begou, H.G. Gika, G.A. Theodoridis, and I.D. Wilson, Methods Mol. Biol. 1738, 15 (2018).
- H.G. Gika, C. Zisi, G. Theodoridis, and I.D. Wilson, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1008, 15 (2016).
- G. Theodoridis, H. Gika, P. Franceschi, L. Caputi, P. Arapitsas, M. Scholz, D. Masuero, R. Wehrens, U. Vrhovsek, and F. Mattivi, Metabolomics 8, 175 (2012).
- O. Begou, H.G. Gika, I.D. Wilson, and G. Theodoridis, Analyst 142, 3079 (2017).
- E.D. Tsochatzis, O. Begou, H.G. Gika, P.D. Karayannakidis, and S. Kalogiannis, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1047, 197 (2017).
- A. Pina, O. Begou, D. Kanelis, H. Gika, S. Kalogiannis, C. Tananaki, G. Theodoridis, and A. Zotou, J. Chromatogr. A 1531, 53 (2018).
- C. Virgiliou, D. Kanelis, A. Pina, H. Gika, C. Tananaki, A. Zotou, and G. Theodoridis, J. Chromatogr. A 1616, 460783 (2020).
- P. Arapitsas, A.D. Corte, H. Gika, L. Narduzzi, F. Mattivi, and G. Theodoridis, Food Chem. 197 Pt B, 1331 (2016).
- O. Begou, O. Deda, A. Agapiou, I. Taitzoglou, H. Gika, and G. Theodoridis, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1114–1115, 76 (2019).
- H.G. Gika, C. Ji, G.A. Theodoridis, F. Michopoulos, N. Kaplowitz, and I.D. Wilson, J Chromatogr A 1259, 128 (2012).
- K. Spagou, G. Theodoridis, I. Wilson, N. Raikos, P. Greaves, R. Edwards, B. Nolan, and M.I. Klapa, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 879, 1467 (2011).
- H.G. Gika, G.A. Theodoridis, and I.D. Wilson, J. Sep. Sci. 31, 1598 (2008).
- P. Pousinis et al., “Comprehensive (Metabolomics and metals) analysis of sow feed,feces and urine: associations with health and welfare indicators”, poster presented at Metabolomics 2021, Online
- N.C. Maragou et al., J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1137, 121938 (2020).
- D. Diamantidou, O. Begou, G. Theodoridis, H. Gika, E. Tsochatzis, S. Kalogiannis, N. Kataiftsi, E. Soufleros, and A. Zotou, J. Chromatogr. A 1603, 165 (2019) .
- E.D. Tsochatzis, H. Gika, and G. Theodoridis, Anal. Chim. Acta 1130, 49 (2020).
Georgios Theodoridis is professor of analytical chemistry at the Department of Chemistry, Aristotle University of Thessaloniki, in Greece. He studied chemistry (1990) and received his Ph.D. (1994) in separations sciences from the Aristotle University. He leads the BIOMIC (Bioanalysis and Omics Interdisciplinary Laboratory) at the Innovation Centre of the Aristotle University (http://biomic.web.auth.gr/) and the FoodOmicsGR_Research Infrastructure.
Petros Pousinis is a chemist with an M.Sc. in analytical chemistry from the Aristotle University of Thessaloniki. He also obtained his Ph.D. degree from the School of Pharmacy at the University of Nottingham, UK, in 2017. He is currently a postdoctoral researcher in the BIOMIC_AUTh group, employing metabolomics methods (both targeted and untargeted LC–HRMS) to study animal health.
Artemis Lioupi obtained her bachelor degree in chemistry from Aristotle University of Thessaloniki in 2017 and her master’s degree in chemical analysis‑bioanalysis in 2019. She is currently a Ph.D. candidate in the Department of Chemistry of Aristotle University of Thessaloniki and a member of the BIOMIC_AuTh team.
Maria Marinaki is a chemical engineer (M.Sc. in the Department of Chemical Engineering, Faculty of Engineering, Aristotle University of Thessaloniki), a PhD candidate in the Department of Chemistry (Laboratory of Analytical Chemistry), and also a member of the BIOMIC Auth Group with a fellowship in the foodomics programme.
Christina Virgiliou studied chemistry at the Aristotle University Thessaloniki (Greece) where she took her PhD on analytical chemistry and specifically on method development for metabolomics-based studies. She is currently technical manager of the BIOMIC laboratory.
Alexandros Pechlivanis is a postdoctoral research associate in the BIOMIC_AUTh group. He obtained his B.Sc in chemistry in 2006 from Aristotle University of Thessaloniki, his M.Sc in 2008 in “analytical chemistry-quality control”,
and his PhD in 2014.
Dritan Kodra is a postdoctoral researcher at the Department of Chemistry, Aristotle University of Thessaloniki (AUTH).
Helen Gika is assistant professor of biomolecular analysis at the Department of Medicine of the Aristotle University of Thessaloniki. She studied chemistry (2000) and received her Ph.D. (2004) in HPLC of hormones from the Aristotle University.
E-mail: gtheodor@chem.auth.gr; petpousinis@hotmail.com; foodomicsgr@gmail.com
Website: http://foodomics.gr/

A Matrix-Matched Semiquantification Method for PFAS in AFFF-Contaminated Soil
Published: April 14th 2025 | Updated: April 14th 2025Catharina Capitain and Melanie Schüßler from the Faculty of Geosciences at the University of Tübingen, Tübingen, Germany describe a novel approach using matrix-matched semiquantification to investigate per- and polyfluoroalkyl substances (PFAS) in contaminated soil.
Silvia Radenkovic on Building Connections in the Scientific Community
April 11th 2025In the second part of our conversation with Silvia Radenkovic, she shares insights into her involvement in scientific organizations and offers advice for young scientists looking to engage more in scientific organizations.













