The Importance of the Sample Matrix
LCGC North America
How to account for the sample matrix when developing a method
It is important to consider the influence of the sample matrix when setting up an analytical method.
When we set up an analytical liquid chromatography (LC) method, our focus is usually on the analyte of interest. However, other things that potentially are present in the sample can have a profound influence on the results we obtain. As a generic term, we use "sample matrix" to describe everything that is present in the typical sample except for the analytes of interest. If we are analyzing an environmental water sample, the matrix would be water without the analyte. For a bioanalytical method intended to measure a drug in plasma, the matrix would be untreated plasma. For a pharmaceutical formulation, it would be a placebo with all the excipients, but no active ingredient. Only if our samples comprise a pure compound, such as with the analysis of a raw material for purity, can we ignore the sample matrix. And in some cases, even a supposedly pure compound may contain other things, such as reaction impurities or by-products. As a result, whenever we are developing or transferring an LC method, it is important to consider what effect the sample matrix may have on the results. If we don't, we may find ourselves in a troubleshooting situation when the results don't make sense.
What Do the Authorities Say?
In the pharmaceutical industry, the regulatory agencies are aware of the importance of the sample matrix. Let's consider a few of these. The International Conference on Harmonization (ICH) publishes guidelines on validation of analytical methods (1). One of the important factors to consider is specificity. The ICH definition states:
Specificity is the ability to assess unequivocally the analyte in the presence of components which may be expected to be present. Typically these might include impurities, degradants, matrix, etc.
You can see that the implication of this definition is that things, other than the analyte, that are present in the sample can confuse the identification or ability to quantify the analyte of interest.
The United States Pharmacopoeia (USP) includes Chapter 1226, "Verification of Compendial Procedures" (2). Although these guidelines technically apply only to USP methods, many workers apply them to non-USP methods, as well. Included in the discussion are suggestions of tests to consider when evaluating methods:
. . . an assessment of specificity is a key parameter in verifying that a compendial procedure is suitable for use in assaying drug substances and drug products . . . drug substances from different suppliers may have different impurity profiles that are not addressed by the compendial test procedure. Similarly, the excipients in a drug product can vary widely among manufacturers and may have the potential to directly interfere with the procedure or cause the formation of impurities that are not addressed by the compendial procedure. In addition, drug products containing different excipients, antioxidants, buffers, or container extractives may affect the recovery of the drug substance from the matrix.
Here again we see that specificity is an important consideration. The USP highlights the possibility that different drug sources may contain different impurities and that different matrices may affect the recovery of the analytes of interest.
A third source of regulatory opinion is seen in advice about the validation of analytical methods to measure drugs present in biological matrices (3) from the United States Food and Drug Administration (FDA). What is referred to as specificity by the ICH and USP is called selectivity by the FDA:
Selectivity is the ability of an analytical method to differentiate and quantify the analyte in the presence of other components in the sample. For selectivity, analyses of blank samples of the appropriate biological matrix (plasma, urine, or other matrix) should be obtained from at least six sources. Each blank sample should be tested for interference, and selectivity should be ensured at the lower limit of quantification (LLOQ).
You can see that the influence of the matrix is important in bioanalytical methods, as well. And, whereas drug product or drug substance matrices are very consistent for a certain product over time, plasma can vary widely from individual to individual, so at least six different sources of blank plasma matrix should be tested for interferences.
Some Scenarios
I've prepared several simulated chromatograms to illustrate the importance of using an appropriate matrix when validating, qualifying, or transferring a method. In Figure 1a, a peak with a retention time of 3.40 min is shown to represent our analyte of interest, injected as a calibration standard in mobile phase as the sample diluent. Figure 1b contains a portion of the chromatogram for the matrix for this particular sample type. There is a large, off-scale peak at 3.00 min and a small matrix peak at 3.60 min. Because there is no analyte in the blank matrix, nothing appears at the expected 3.40 min retention time of the analyte. When we spike the blank matrix with the analyte, we get the results shown in Figure 1c. This simply is a combination of the previous two chromatograms. Although it looks like there is baseline separation between the two small peaks, the resolution, Rs, is only 1.43. Usually we would like a minimum resolution of 1.7, or better Rs ≥ 2, for complete separation. And remember that these are ideal, Gaussian-shaped peaks; real peaks with even a minor amount of tailing would have a worse separation. However, in spite of the lack of baseline resolution, the area of the analyte should be easy to determine, if the retention times are steady from run to run, so interferences from the matrix should be a minor concern in this case.
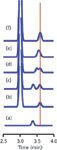
Figure 1: Simulated chromatograms to demonstrate method specificity: (a) analyte only, (b) matrix blank only, (c) matrix spiked with analyte under conditions of (b), (dâf) sample of (c) with different retention for analyte (see text for details).
The situation gets a bit more complicated in the chromatogram of Figure 1d, where I've shifted the retention time of the analyte to 3.50 min. Now the analyte peak overlaps slightly with the minor peak in the matrix, with Rs = 0.70. The overlap is sufficient that it is not possible to accurately measure the peak area of either of these small peaks, but there is enough separation to confidently determine if one or both peaks are present in a given sample.
The problem becomes more acute when resolution is reduced even further. In Figure 1e, I've assigned a retention time of 3.55 min to the analyte, which drops resolution to 0.35. We can no longer tell if a second peak is present. The peaks overlap sufficiently that there is no clear apex for the individual peaks, as we saw in the previous examples. Note that the apparent retention time of the peak that appears is shifted slightly (I've added a vertical line through the chromatograms at 3.60 min as a reference point). Besides the shift in apparent retention time, the combined peak is wider than either of the small peaks when they don't overlap. You might think that you could be alerted to the presence of two peaks by using the peak-purity function that is part of the software of most diode-array ultraviolet detection (DAD) systems. If the UV spectra are sufficiently different, this may be possible. However, although I have seen plenty of examples where this is the case under ideal circumstances, most users find that DAD peak purity is of marginal use in many cases. This is because if the peaks are this closely eluted, it is likely that they are similar compounds and thus have similar spectra. As the signal-to-noise ratio decreases, the spectra become more similar, and the peaks move closer together, the peak purity algorithms tend to have problems. You also might guess that two peaks were present because of the broader peak, but sometimes peak width differences are difficult to measure when the peaks are small.
The final case shown in Figure 1f has the analyte peak at the same retention time as the small interference, 3.60 min. Now it is not possible to tell whether the response is for the analyte, the interference, or a combination of the two. If you were using mass spectrometry (MS) detection, you might be able to measure the two compounds independently, but only if they had different mass-to-charge ratios and there was no ion suppression of one peak by the other. Sometimes other detectors might be able to tell how much response belonged to each peak. For example, if the UV or fluorescence spectra were significantly different, you might be able to measure each peak independently. In any case, this degree of overlap is likely to compromise the results of the analysis. Sometimes with overlapped peaks it is possible to tell if a peak is impure, but it is practically impossible to tell with 100% certainty if a peak is pure.
The examples shown in Figure 1 are idealized, with perfectly shaped peaks, a stable baseline, good signal-to-noise ratios, and similar-sized peaks. The situation gets much more challenging when the chromatogram is more complex. When the peak heights are different, one or both peaks tail, the baseline drifts and is noisy, and other more realistic conditions affect the separation, the ability to be confident of method specificity lies mainly in separating the peaks in time to be sure there is no overlap.
What Matrix to Choose
At the beginning of this discussion, we saw that the various regulatory agencies agreed that specificity should be demonstrated in the presence of the other chemicals that are likely to be present in the sample. Also, the calibration standards should be prepared in a blank matrix, because sometimes the matrix enhances the analyte signal and other times it depresses it. So, what do you choose for a matrix?
If your sample is a formulated product, such as a drug product or a pesticide formulation, selecting the matrix is straightforward. Just use a combination of all the excipients and other ingredients that will be in the product, but leave out the analyte of interest. In other cases, the matrix may be quite simple and you can substitute a similar matrix from a different source. For example, if the method is for the analysis of a pesticide in river water, you may be able to find another water source that you know is untreated, but otherwise very similar.
In other cases, the matrix may be quite complex and not fully under your control. In the FDA's guidelines for the analysis of drugs in biological matrices noted above (3), it is suggested that untreated matrix from six sources be checked for potential interferences. For example, it is common to check six lots of plasma to see if any peaks appear at the same retention time as the analyte of interest. But even this is not foolproof. I remember one LC–MS method in our laboratory in which the method was developed and checked in this manner. It was only when we received samples from treated subjects that we discovered there was an interference problem. How did we miss this? Our plasma sources were those typically purchased from commercial sources. These usually are obtained from healthy volunteers, such as college students who want to make a little extra money by selling their blood. However, we found that the clinical study was run in an Asian country, so the genetics and diet were different, plus the subjects were all terminal cancer patients. These various factors resulted in a quite different chromatographic profile of the blank plasma matrix. The take-home lesson here is that you should try to obtain a blank matrix from a source that is as close as possible to the final samples. In this case, if we could have obtained a plasma sample from a predosed subject, we would have been much more likely to discover the lack of specificity of the method.
In still other cases, it may not be possible to obtain a true blank matrix. For example, if the method is meant to measure naturally occurring compounds in plant tissue or trace by-products in a chemical reaction mixture, it may be impossible to obtain a sample with no analyte present. In such cases, you have to be creative and do your best to create a blank or analyte-depleted matrix to help you show specificity. Techniques such as running the "blank" and then spiking known amounts of analyte, or using LC–MS techniques may help assure you that nothing is present that will compromise the analysis of the analyte because of a lack of method specificity.
Summary
We have seen that when developing, validating, or transferring an LC method it is important to demonstrate that the measurement of the analyte of interest is not compromised by the presence of some other compound in the sample. This is important enough that regulatory guidelines prominently mention specificity as a key component of testing for reliable analytical methods. Sometimes a blank matrix is simple to obtain, but in other cases, sample-to-sample variations in the matrix must be tested on a reasonable number of matrix sources. You also should test for potential interferences from other compounds that may or may not be present in the sample, such as co-dosed drugs or over-the-counter medications. Although sophisticated LC detectors may help to deconvolute partially overlapped peaks, the best choice always is to achieve chromatographic separation between the analyte and any potential interferences.
References
(1) International Conference on Harmonization, Validation of Analytical Procedures: Text and Methodology, Q2(R1), (ICH, Geneva, Switzerland, November 2005).
(2) General Chapter 1226 "Verification of Compendial Procedures," in United States Pharmacopeia 35–National Formulary 30, (United States Pharmacopeial Convention, Rockville, MD, 2012).
(3) US Food and Drug Administration, Guidance for Industry: Bioanalytical Method Validation, (FDA, Rockville, MD, May 2001).

John W. Dolan
John W. Dolan "LC Troubleshooting" Editor John Dolan has been writing "LC Troubleshooting" for LCGC for more than 25 years. One of the industry's most respected professionals, John is currently the Vice President of and a principal instructor for LC Resources, Walnut Creek, California. He is also a member of LCGC's editorial advisory board. Direct correspondence about this column via e-mail to John.Dolan@LCResources.com

Extracting Estrogenic Hormones Using Rotating Disk and Modified Clays
April 14th 2025University of Caldas and University of Chile researchers extracted estrogenic hormones from wastewater samples using rotating disk sorption extraction. After extraction, the concentrated analytes were measured using liquid chromatography coupled with photodiode array detection (HPLC-PDA).
Polysorbate Quantification and Degradation Analysis via LC and Charged Aerosol Detection
April 9th 2025Scientists from ThermoFisher Scientific published a review article in the Journal of Chromatography A that provided an overview of HPLC analysis using charged aerosol detection can help with polysorbate quantification.
Removing Double-Stranded RNA Impurities Using Chromatography
April 8th 2025Researchers from Agency for Science, Technology and Research in Singapore recently published a review article exploring how chromatography can be used to remove double-stranded RNA impurities during mRNA therapeutics production.








