Identifying Differences in Solution Conformations of Two Chimeric IgG3 Antibodies through Triple Detection SEC
LCGC Europe
Size exclusion chromatography (SEC) coupled to light-scattering, viscometer and refractive index detectors is a common technique used for the characterization of polymers. In this article the solution conformations of two chimeric IgG3 antibodies were studied using this coupled triple detection technique. Conclusions indicate that the results allow hydrodynamic modelling of the antibody structures.
Introduction
By identifying the change in viscosity of a solution upon addition of a protein, a parameter known as the intrinsic viscosity can be determined for that protein. The intrinsic viscosity of a protein is a value partially dependent on protein conformation. From the intrinsic viscosity value, a viscosity increment value can be determined, which is a size-independent value dependent solely upon the shape of a protein. The higher the viscosity increment, the more extended is the protein that it represents.

Antibody molecules are proteins, which play vital roles in the body's immune system. They can be split into five classes: IgM, IgG, IgA, IgD and IgE which, although they each have unique structural and functional properties, all have the same basic structure. Some of these classes of antibodies can be further sub-divided into subclasses. The IgG class, for example, consists of four subclasses: IgG1, IgG2, IgG3 and IgG4.
The subclass IgG3 antibodies were investigated in this study because of their unusually long "hinge" region that gives this antibody the extra flexibility that is vital for its function.
The two chimeric IgG3 molecules investigated in this study were a wild type IgG3 with the full length 62 amino acid hinge [Figure 1(a)], and a hinge-deleted mutant IgG3 with a 15 amino acid hinge [Figure 1(b)], developed by Michaelsen and coworkers.1
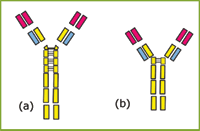
Figure 1: Schematic diagram of (a) IgG3wt and (b) IgG3m15. Two 15 amino acid segments and one 17 amino acid segment, present in the hinge of IgG3wt, are absent in the hinge of IgG3m15.
These antibodies were developed to aid understanding of the functional significance of the extended IgG3 hinge. This study aimed to identify the conformational differences caused by this hinge elongation, and relate these differences to their differing abilities to activate the complement cascade.
High-resolution techniques such as x-ray crystallography and nuclear magnetic resonance (NMR) are commonly used for protein structure determination. Studies of the solution conformations of antibodies have proved complicated, however, with NMR having an upper molecular weight cut off of ~50000 Da, whilst IgG3 has a molecular weight of ~160000 Da. Additionally, crystallographic structures of intact IgG molecules tend to have poorly characterized diffraction patterns around the hinge and Fc region of the molecule because of the flexibility of the molecules.
An alternative approach to studying antibody conformation is to look at hydrodynamic parameters. The intrinsic viscosity parameter was chosen to investigate the solution conformations of IgG3 molecules in this study. It was hoped that by obtaining the intrinsic viscosity values of the two antibodies and subsequently creating models based on this shape information, conclusions could be drawn on the differences in their solution conformations. An advantage of using a solution technique is that models represent the structure of the protein in a dilute solution, which is more likely to relate to its conformation in vivo.
Background
Combining size exclusion chromatography (SEC) with light-scattering (LS), viscometer and refractive index (RI) detectors is a powerful tool for protein characterization. This approach provides not only molecular weights (M) and the intrinsic viscosity ([η]) but also quantifies aggregates, determines hydrodynamic radii (Rh) and provides conformation information from a single SEC run. The advantage of this combination of detectors is that the sample properties can be derived directly from the detector signals. The signals of the individual detectors correspond to
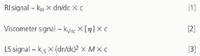
[η] = intrinsic viscosity
c = concentration
dn/dc = refractive index increment, kRI, Visc, LS = detector constants
The refractive index detector determines the refractive index increment, which is necessary for the calculation of the molecular weight from the LS signal. The intrinsic viscosity is obtained from the viscometer signal after the concentration is appropriately taken into consideration.2
The intrinsic viscosity of a protein is a function of shape, hydration and flexibility. If it is assumed that the hydration of an antibody is known, the shape information that can be derived from an intrinsic viscosity value can be used to model the solution conformation of a protein.

The partial specific volume is the anhydrous volume per unit mass of a molecule dissolved in a solution. The viscosity increment is a "universal shape parameter",4,5 meaning its value is dependent solely on shape.
Additionally from molecular weight and intrinsic viscosity, the hydrodynamic radius Rh of the protein can be obtained. This parameter is determined using Einstein's equation for hard spheres,6,7 which states that hydrodynamic volume (Vh) is proportional to the product of molecular weight and intrinsic viscosity (Equation 5). The hydrodynamic radius is obtained from a combination of Vh and Equation 5, as shown in Equation 6.9
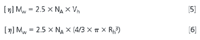
NA= Avogadro's number
Rh depends strongly on shape and this parameter can be used to differentiate between spherical or compact proteins and more elongated proteins.
Modelling of Antibody Structure
Universal shape parameters such as the viscosity increment can be used to model the conformation parameters. Previous studies8, 9 have determined solution conformations of antibodies using a universal shape parameter obtained from the sedimentation coefficient — the Perrin function. By representing the Fab and Fc domains as ellipsoids and altering the antibody inter-domain angles, agreement was found between the experimental Perrin functions of the proteins and the theoretical Perrin functions from the models.
The same approach can be applied to modelling based on the viscosity increment, to find the solution conformations of the two IgG3 antibodies.
Materials and Methods
The chromatographic system consisted of a Viscotek GPCmax (degasser, pump and automatic sampler with 100 μL sample loop) and a Viscotek TDA 302 (containing differential refractive index, light scattering and viscometer detectors). Figure 2 shows how the output of the GPCmax is connected to the inlet of the light scattering detector, refractive index detector (RI) and a four-capillary differential viscometer detector. The series configuration is important as it yields the maximum signals on all three detectors.
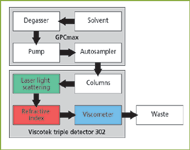
Figure 2: Diagram of the GPCmax and the triple detection array.
The separation of the IgG samples were performed on two Tosoh columns (G3000SWxl and G4000SWxl; 7.8 mm × 30 cm) using phosphate buffered saline (PBS) buffer as mobile phase at a flow rate of 0.7 mL/min. The detector and columns were equilibrated at a temperature of 26 °C. The PBS buffer was filtered with a 0.2 μm nylon membrane filter. The IgG samples were also prepared in PBS buffer.
For the Viscotek triple detector calibration a pullulan Standard of known concentration, molecular weight, intrinsic viscosity and refractive index increment (dn/dc) was used for determining the instrument constants of the three detectors. The method was subsequently validated with a second reference standard (Dextran T70).
The instrument control, data acquisition and all calculations were performed using OmniSEC software (Viscotek, version 2.0).
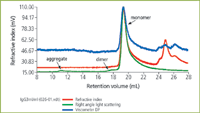
Figure 3: Triple detection chromatogram of IgG3m15 (mutant type).
Results and Discussion
The objective of this study was to investigate the conformations of the two IgG3 proteins.
The chromatograms for the two proteins IgG3m15 (mutant type) and IgGwt (wild type) are shown in Figures 3 and 4 respectively. Both antibodies give clear monomer peaks, IgG3m15 at a 19.5 mL retention volume and IgG3wt at 18.5 mL. The presence of dimer was detected in the IgG3wt sample at a retention volume of 16.5 mL. This result was consistent with results from sedimentation velocity experiments (data not shown). The presence of dimer and higher aggregates were detected by light scattering in the IgG3m15 sample at retention volumes of 17.5 mL and 11 mL respectively. These results reveal the power of LS measurements in the detection of trace amounts of aggregate in protein samples.
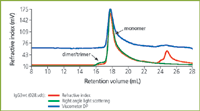
Figure 4: Triple detection chromatogram of IgG3wt (wild type).
Table 1 shows the results obtained for the two IgG3 antibodies. Since the objective in this study was to compare the relative changes in molecular properties a typical protein dn/dc value of 0.185 mL/g for IgG was used in all calculations. The determination of "absolute" molecular weight requires a precise knowledge of the dn/dc or an accurate concentration of the protein, with no artefacts caused by interaction of protein with column matrix. In this work it can clearly be seen from the chromatograms that there is some interaction of the antibodies with the column matrix, causing tailing of the detector signals. Of course optimizing run conditions could minimize the column interaction.

Table 1: Results determined through triple detection of IgG3wt and IgG3m15 antibodies.
A very significant difference is observed between the viscosity values obtained for the two IgG3 proteins. The extended wild-type IgG3, IgG3wt, has an intrinsic viscosity of 9.9 mL/g compared with the much lower value of 5.7 mL/g for the hinge-deleted mutant, IgG3m15. The value for IgG3m15 is similar to the value of 6.2 mL/g obtained previously by Kilar and coworkers10 for an IgG1 antibody with a similar length hinge. A value for hydration of 0.59 mL/g was used, as determined by Carassco et al.8 These intrinsic viscosity values, when converted to viscosity increments, give values of 7.5 and 4.3 for the IgG3wt and IgG3m15 respectively.

Figure 5: Hydrodynamic model representing the conformation of IgG3wt (wild type). Fab and Fc domains are represented by hydrodynamically equivalent ellipsoids. The hinge is represented by frictionless connectors.
A high viscosity increment is an indication of an extended molecule and, therefore, the higher value for the IgG3wt protein reveals that this antibody is more elongated than IgG3m15. This is also indicated by the IgG3wt antibody having a higher Rh of 6.4 nm, relative to the value of 5.1 nm for IgG3m15.
Figures 5 and 6 show examples of size-independent models which are hydrodynamically equivalent to the IgG3wt and IgG3m15 antibodies, on the basis of their viscosity increment values. It is clear that the model representing the potential conformation of IgG3wt is significantly more extended than the model for IgG3m15.

Figure 6: Hydrodynamic model representing the conformation of IgG3m15 (mutant type). Fab and Fc domains are represented by hydrodynamically equivalent ellipsoids. The hinge is represented by frictionless connectors.
Conclusions
Triple detection has been shown to be highly effective for the analysis of the solution properties of the antibodies. This approach has not only provided quantitative information on self-association present in the samples, but also provided conformation information of sufficient resolution to allow hydrodynamic modelling of protein conformation. On the basis of this information it was possible to deduce that the partial deletion of the hinge of an IgG3 antibody (IgG3wt) to produce the hinge deleted mutant (IgG3m15), resulted in an antibody with a significantly more compact solution conformation. For the wild type IgG3 antibody, IgG3wt, with an extended hinge region, a viscosity increment value of 7.5 was obtained. The less extended hinge-deleted mutant IgG3 antibody, IgG3m15, gave a viscosity increment of 4.3, consistent with this being a more compact protein.
References
1. T.E. Michaelsen et al., Scandinavian Journal of Immunology, 32, 517–528 (1990).
2. S.E. Harding, Progress in Biophysics & Molecular Biology, 68, 207–262 (1997).
3. J.T. Yang, Advances in Protein Chemistry, 16, 323–400 (1961).
4. J. Garcia de la Torre, European Biophysics Journal with Biophysics Letters, 25, 361–372 (1997).
5. S.E Harding et al., European Biophysics Journal with Biophysics Letters, 25, 347–359 (1997).
6. A. Einstein, Annalen der Physik (Leipzig), 19, 289–305 (1906).
7. A. Einstein, Annalen der Physik (Leipzig), 34, 591–593 (1911).
8. B. Carrasco et al., Biophysical Chemistry, 93, 181–196 (2001).
9. E. Longman et al., European Biophysics Journal with Biophysics Letters 32, 503–510 (2003).
10. F. Kilar, European Journal of Biochemistry, 147, 17–25 (1985).
Emma Longman undertook the research on the solution conformations of antibodies for her PhD at the National Centre for Macromolecular Hydrodynamics at the University of Nottingham, UK. She now works as a postdoctoral research fellow in the Biophysics group of the Structural Genomics Consortium at the University of Oxford, UK. Nicola Marheineke obtained her degree in Chemistry at Europa Fachhochschule Fresenius in Idstein, Germany. She worked for over six years as a GPC/SEC application chemist with Viscotek Europe and is now currently studying at the University of Pierre et Marie Curie, Paris, France. Stephen Harding received a DSc degree from the University of Oxford, UK. He is professor of physical biochemistry and director of the national centre for macromolecular hydrodynamics at the University of Nottingham.

Understanding FDA Recommendations for N-Nitrosamine Impurity Levels
April 17th 2025We spoke with Josh Hoerner, general manager of Purisys, which specializes in a small volume custom synthesis and specialized controlled substance manufacturing, to gain his perspective on FDA’s recommendations for acceptable intake limits for N-nitrosamine impurities.









