How Much Data is Too Much? An Analysis of the Pros and Cons of High-Resolution Mass Spectral Data
The introduction of high-resolution mass spectrometry (HRMS) to the field of chromatography has significantly increased the amount of information gained from analysis of complex samples. HRMS yields results with higher mass accuracy than previously possible, but this ability to precisely measure masses presents the problem of applying chemometric analyses to an exponentially larger dataset. This article will evaluate the pros and cons of evaluating HRMS data for the detection and identification of analytes present in complex mixtures.
As the instrumentation in the separation science field improves, the amount of chemical information that can be gained has exponentially increased. One of the best examples is the introduction of high-resolution mass spectrometry (HRMS) to gas chromatography (GC) and comprehensive two-dimensional gas chromatography (GC×GC) (1–12). Commercially-fielded high-resolution mass spectrometers, such as orbital trap and time-of-flight mass spectrometers, can measure masses accurately down to the sixth and fourth decimal place, respectively (13–16). With these mass spectrometers comes new software that allows for data analysis such as Kendrick Mass Defect (KMD), Van Krevelen, and Ring Double Bond Equivalency (RDBE) based on carbon number. This presents a major improvement for spectral analysis. While these methods provide improved spectral analysis, the application of HRMS also allows for unique application of external chemometric software for the identification of non-targeted compounds. However, a downside to HRMS is the large data file sizes, which are difficult to manipulate and evaluate in statistical software such as R, MATLAB, or Python.
A recent study performed at Los Alamos National Laboratory analyzed a set of complex high vacuum pump oils using comprehensive two-dimensional gas chromatography with high-resolution time-of-flight mass spectrometry (GC×GC–HRMS). The method used for this experiment was derived and modified from a webinar given by Gröger and associates in 2016 (17). In this experiment, an Agilent 8890 coupled to a LECO Pegasus HRT+ (LECO) was utilized to analyze the samples in electron ionization and negative and positive chemical ionization. Due to the high boiling point compounds found within high-vacuum pump oils, a large temperature range was necessary for analysis. Two high-temperature columns used GC×GC with the first dimension being a ZB-35 HT 30 m × 0.25 mm × 0.25 μm (Phenomenex), and the second dimension being ZB-1HT 4 m × 0.18 mm × 0.18 μm (Phenomenex). The oven was held at 175 °C for 1 min, and then ramped at 1 °C/min up to 340 °C. This temperature programming results in a runtime of over three hours!
While the runtime is a major contributor to how large the datasets are, high-resolution mass spectrometry exponentially increases the number of data points in a single sample. Mass spectra from 50 to 600 amu to the nearest 0.0001 amu were collected at a rate of 100 Hz. This resulted in a possible 5.5 million data points for each mass spectral scan.
When multiplying the number of mass channels by the number of scans over the runtime for each sample, there is a possibility of having trillions of data points per sample. Though not every mass channel is present at every scan, a single chromatographic run after being exported as a .csv from ChromaTOF (Version 5.53.70.0.113) software is still over 30 GB (Figure 1)! Exporting data files of this size can take long periods of time, and although the export process was automated via the ChromaTOF software, each file required 10 min to be compiled into .csv files.
FIGURE 1: Schematics of GC×GC–HRMS data exportation and chemometric analysis of large sample datasets.
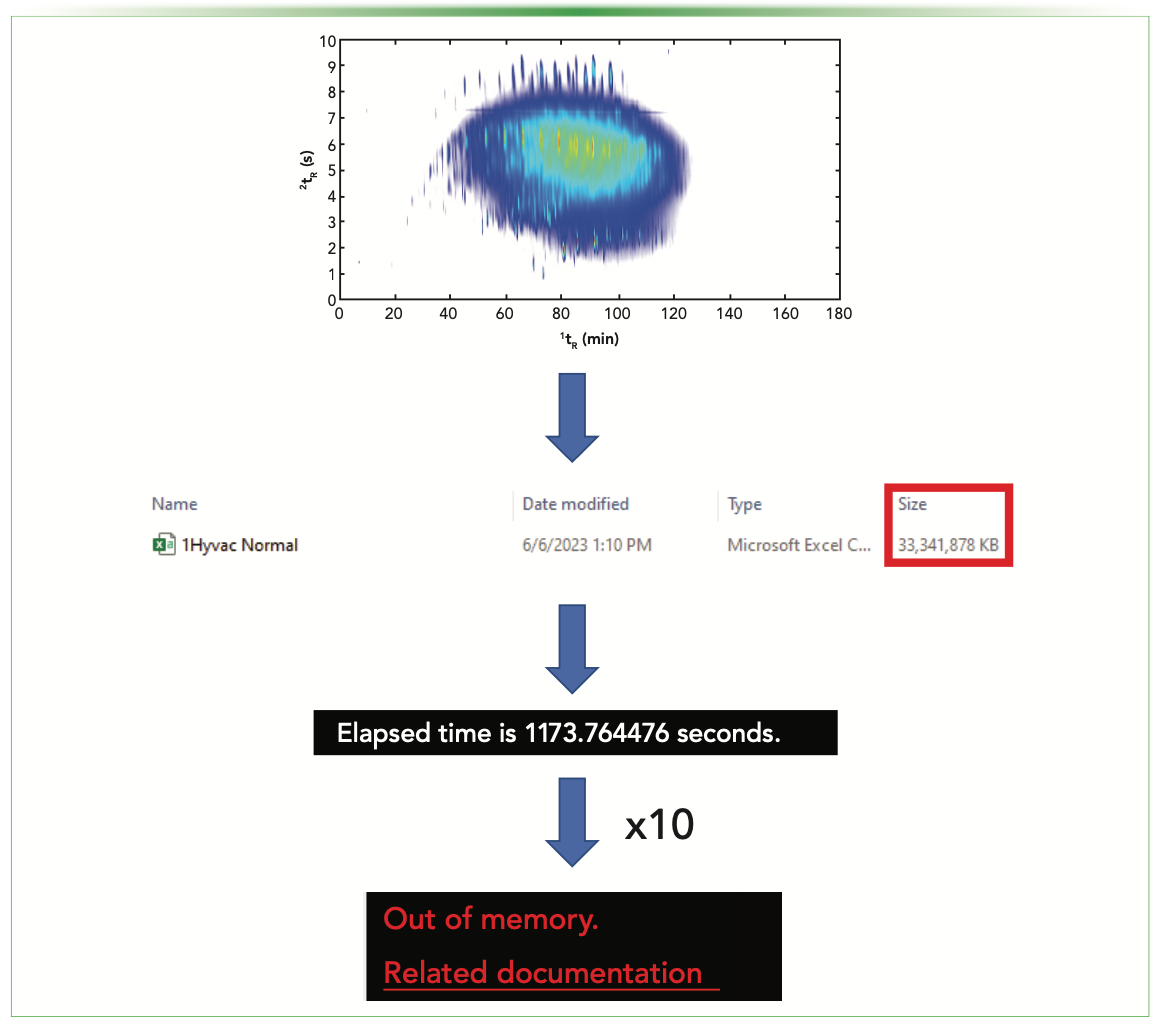
Once the data was exported, the samples were statistically evaluated using MATLAB 2022a (Mathworks). Because of the large data sizes, it took ~17 min to import each sample into MATLAB. To chemometrically evaluate the samples, multiple data files needed to be uploaded and stored in the global environment to further undergo data reduction. However, after approximately 10 sam- ples were uploaded, the code returned the error, “Out of memory” (Figure 1).
Running out of memory on a computer is a difficult problem to solve. The few options available to a chemometrician when running out of memory include buying a new computer with more RAM (potentially a cluster), or binning the data that was collected to a level where the datasets are smaller. The second option is typically what scientists will settle on, but this required taking the collected data for the high-vacuum pump oils and binning the mass spectra into unit resolution. From the unit resolution data there is a possibility of analyzing these large data files, and if regions of interest are found in specific portions of the sample, a scientist can go back and analyze the HRMS data of these regions. However, this nullifies the use of HRMS data in multiple regions of the sample where areas of interest may be present but are not being identified due to a simplification of the data.
This case leads to the question: “How do we fix this?” The answer is not simple, and could potentially require improvements in computer data storage for samples with long runtimes to be analyzed using a technique like HRMS. Similarly, more complex computing power, including clusters or supercomputers, may be required in the present day for analysis of big data at this scale. While this may not be available to the typical chromatographer, it may be necessary when trying to analyze and characterize samples that are complex in nature, such as higher boiling point petroleum products, especially when analyzed using GC×GC.
A paper by Thompson and associates explains that improvements to hardware in computers are currently slowing down, which will constrain the use of improved chemometric techniques, mainly deep learning, and require the use of more computationally efficient machine learning techniques (18). To improve classifications and comparisons that can be made with big data, machine learning techniques that are more memory-efficient need to be explored. Hopefully, in the future, hardware improvements will allow for more deep learning methodologies to be applied to big data, such as GC×GC– HRMS data.
References
(1) Idowu, I.; Johnson, W.; Francisco, O.; Obal, T.; Marvin, C.; Thomas, P. J.; Sandau, C. D.; Stetefeld, J.; Tomy, G. T. Comprehensive Two-Dimensional Gas Chromatography High-Resolution Mass Spectrometry for the Analysis of Substituted and Unsubstituted Polycyclic Aromatic Compounds in Environmental Samples. J Chromatogr A 2018, 1579, 106–114. DOI: 10.1016/j.chroma.2018.10.030
(2) Byer, J. D.; Siek, K.; Jobst, K. Distinguishing the C3 vs SH4 Mass Split by Comprehensive Two-Dimensional Gas Chromatography–High Resolution Time-of-Flight Mass Spectrometry. Anal. Chem. 2016, 88 (12), 6101–6104. DOI: 10.1021/acs.analchem.6b01137
(3) Randazzo, G. M.; Bileck, A.; Danani, A.; Vogt, B.; Groessl, M. Steroid Identification via Deep Learning Retention Time Predictions and Two-Dimensional Gas Chromatography-High Resolution Mass Spectrometry. J. Chromatogr. A 2020, 1612, 460661. DOI: 10.1016/j.chroma.2019.460661
(4) Schwalb, L.; Tiemann, O.; Käfer, U.; Gröger, T.; Rüger, C. P.; Gayko, G.; Zimmermann, R. Analysis of Complex Drugs by Comprehensive Two-Dimensional Gas Chromatography and High-Resolution Mass Spectrometry: Detailed Chemical Description of the Active Pharmaceutical Ingredient Sodium Bituminosulfonate and its Process Intermediates. Anal. Bioanal. Chem. 2023, 415 (13), 2471–2481. DOI: 10.1007/s00216-022-04393-w
(5) Alaee, M.; Sergeant, D. B.; Ikonomou, M. G.; Luross, J. M. A Gas Chromatography/High-Resolution Mass Spectrometry (GC/HRMS) Method for Determination of Polybrominated Diphenyl Ethers in Fish. Chemosphere 2001, 44 (6), 1489–1495. DOI: 10.1016/s0045-6535(00)00311-8
(6) Špánik, I.; Machyňáková, A. Recent Applications of Gas Chromatography with High-Resolution Mass Spectrometry. J. Sep. Sci. 2018, 41 (1), 163–179. DOI: 10.1002/jssc.201701016
(7) Samanipour, S.; Langford, K.; Reid, M. J.; Thomas, K. V. A Two Stage Algorithm for Target and Suspect Analysis of Produced Water via Gas Chromatography Coupled with High Resolution Time of Flight Mass Spectrometry. J. Chromatogr. A 2016, 1463, 153–161. DOI: 10.1016/j.chroma.2016.07.076
(8) Peterson, A. C.; Balloon, A. J.; Westphall, M. S.; Coon, J. J. Development of a GC/Quadrupole-Orbitrap Mass Spectrometer, Part II: New Approaches for Discovery Metabolomics. Anal. Chem. 2014, 86 (20), 10044–10051. DOI: 10.1021/ac5014755
(9) Peterson, A. C.; Hauschild, J.-P.; Quarmby, S. T.; Krumwiede, D.; Lange, O.; Lemke, R. A.; Grosse-Coosmann, F.; Horning, S.; Donohue, T. J.; Westphall, M. S.; Coon, J. J.; Griep-Raming, J. Development of a GC/Quadrupole-Orbitrap Mass Spectrometer, Part I: Design and Characterization. Anal. Chem. 2014, 86 (20), 10036–10043. DOI: 10.1021/ac5014767
(10) Sampat, A. A.; Lopatka, M.; Vivó-Truyols, G.; Schoenmakers, P. J.; van Asten, A. C. Towards Chemical Profiling of Ignitable Liquids with Comprehensive Two-Dimensional Gas Chromatography: Exploring Forensic Application to Neat White Spirits. Forensic Sci. Int. 2016, 267, 183–195. DOI: 10.1016/j.forsciint.2016.08.006
(11) Shevyrin, V.; Melkozerov, V.; Nevero, A.; Eltsov, O.; Morzherin, Y.; Shafran, Y. 3-Naphthoylindazoles and 2-naphthoylbenzoimidazoles as Novel Chemical Groups of Synthetic Cannabinoids: Chemical Structure Elucidation, Analytical Characteristics and Identification of the First Representatives in Smoke Mixtures. Forensic Sci. Intl. 2014, 242, 72–80. DOI: 10.1016/j.forsciint.2014.06.022
(12) Stefanuto, P.-H.; Focant, J.-F. GC× GC-TOFMS, the Swiss Knife for VOC Mixtures Analysis in Soil Forensic Investigations. In Soil in Criminal and Environmental Forensics: Proceedings of the Soil Forensics Special, 6th European Academy of Forensic Science Conference, The Hague, 2016; Springer: pp 317–329.
(13) Marshall, A. G.; Hendrickson, C. L. High-Resolution Mass Spectrometers. Annu. Rev. Anal. Chem. 2008, 1 (1), 579–599. DOI: 10.1146/annurev.anchem.1.031207.112945
(14) Xian, F.; Hendrickson, C. L.; Marshall, A. G. High Resolution Mass Spectrometry. Anal. Chem. 2012, 84 (2), 708–719. DOI: 10.1021/ac203191t
(15) Kaufmann, A. The Current Role of High-Resolution Mass Spectrometry in Food Analysis. Anal. Bioanal. Chem. 2012, 403 (5), 1233–1249. DOI: 10.1007/s00216-011-5629-4
(16) Hernández, F.; Portolés, T.; Pitarch, E.; López, F. J. Gas Chromatography Coupled to High-Resolution Time-of-Flight Mass Spectrometry to Analyze Trace-Level Organic Compounds in the Environment, Food Safety and Toxicology. Trends Analyt. Chem. 2011, 30 (2), 388–400. DOI: 10.1016/j.trac.2010.11.007
(17) Käfer, U.; Jennerwein, M.; Weggler, B.; Eschner, M.; Zimmermann, R.; Gröger, T. High Temp GC×GC of Light Crude Oil and High Boilers Using Nominal and High Resolution TOF MS. LECO 2016 https://vimeo.com/259202333/5bf2fa7795
(18) Thompson, N. C.; Greenewald, K.; Lee, K.; Manso, G. F. The Computational Limits of Deep Learning. arXiv:2007.05558v2. DOI: 10.48550/arXiv.2007.05558
Michelle Corbally is a Postdoctoral Researcher in the High Explosives Science & Technology group at Los Alamos National Laboratory, in Los Alamos, New Mexico. Chris Freye is a Scientist in the High Explosives Science & Technology group at Los Alamos National Laboratory, in Los Alamos, New Mexico. Direct correspondence to: mcorbally@lanl.gov

New Method Explored for the Detection of CECs in Crops Irrigated with Contaminated Water
April 30th 2025This new study presents a validated QuEChERS–LC-MS/MS method for detecting eight persistent, mobile, and toxic substances in escarole, tomatoes, and tomato leaves irrigated with contaminated water.
University of Tasmania Researchers Explore Haloacetic Acid Determiniation in Water with capLC–MS
April 29th 2025Haloacetic acid detection has become important when analyzing drinking and swimming pool water. University of Tasmania researchers have begun applying capillary liquid chromatography as a means of detecting these substances.
Prioritizing Non-Target Screening in LC–HRMS Environmental Sample Analysis
April 28th 2025When analyzing samples using liquid chromatography–high-resolution mass spectrometry, there are various ways the processes can be improved. Researchers created new methods for prioritizing these strategies.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)







