Functional Copolymers Analysis Using Liquid Chromatography (LC)—Sort It Out
Polymers are becoming more and more important to our society, with very broad applications ranging from automotive to electronics, textiles, con‑ struction, packaging, and medical use. The main drivers of new material development are twofold. One driver is the development of higher performing functional polymers with special structural features. These can be very specific properties tailored to the desired application—for example, drug release, barrier, or degradation profiles. The second driver is an aim to improve sustainability by reusing materials to reduce their carbon footprints. Examples include bio‑based and environmentally friendly materials, life cycle management, and microplastic reduction. Moreover, current polymer research will help to solve current global sustainability challenges. These two strong driving forces for innovation have resulted in a dramatic increase in the overall complexity of their molecular structure—and their interrelated distributions. Indeed, copolymers composed of much more than three different monomers or oligomers are no longer exceptional. As a result, the complexity of the copolymers increases the dimensionality of the sample, featuring interrelating distributions in molar mass (MWD), chemical composition (CCD), block length (BLD), functionality type (FTD), molecular architecture (MAD), and branching (BD). Consequently, a thorough understanding of the relation between molecular structure, including interrelating distributions and final properties, is a crucial aspect in current polymer research and in innovation towards new, sustainable, and functional copolymers. To achieve this, a well-developed analytical toolbox of high-end methodologies and technologies is needed to elucidate the molecular structures of copolymers, and to develop a thorough understanding of the relationship between their molecular distribution structures. These insights will enhance the transition into new sustainable functional copolymers.
3D image of Pentylene Glycol skeletal formula - molecular chemical structure of Methylethylene glycol isolated on white background | Image Credit: © kseniyaomega - stock.adobe.com
Liquid chromatography (LC) is an important technology to help determine the molecular structure and distribution of copolymers. Due to the increasing complex‑ ity of these copolymers, generation of mono‑distribution information— where each present chemical-feature distribution is fully separated from oth‑ ers—is key here. Recent developments in LC in the areas of sample preparation, mono‑distribution separations, detec‑ tion, data analysis, and hyphenation into fully orthogonal and optimized two‑dimensional chromatography (2DLC) separations have unlocked the ability to achieve insights into monodistribution information and their inter‑ relating distributions (1).
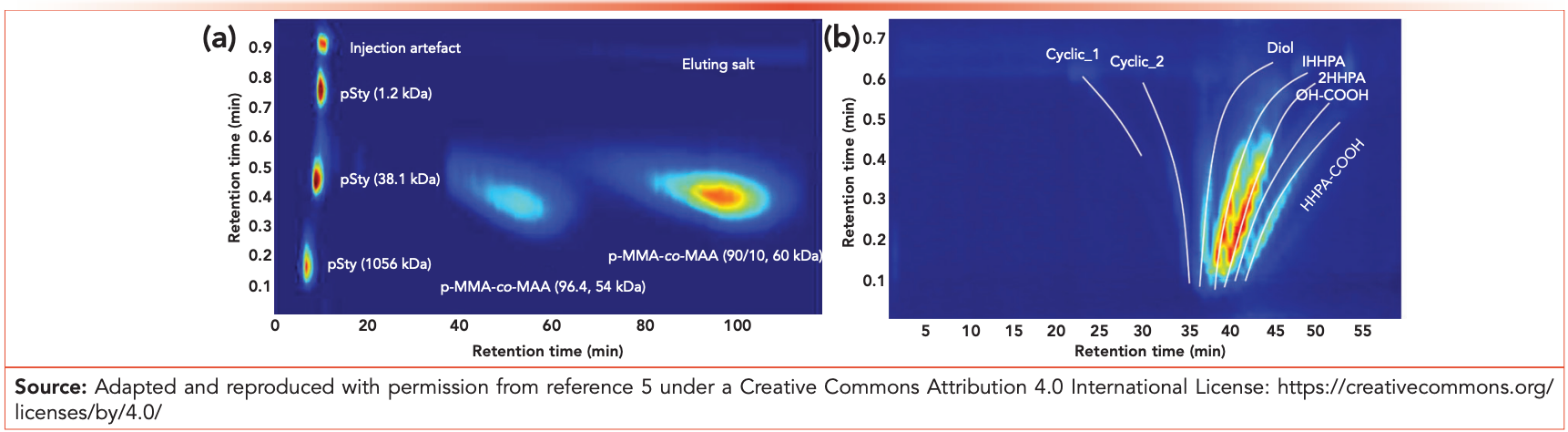
Mono-distributions include, for example, particle size, chemical composition, functionality, and sequence length distributions over the molecular weight distribution of these functional polymers. In the last few years, several mono-distribution separations have been devel‑ oped. One example is the impossible online coupling of hydrodynamic chromatography (HDC) with size-exclusion chromatography (SEC), resulting in the particle size distribution of polymeric nanoparticles over the molecular weight of the constituent polymers (2). This resulted in new insights into how polymeric nanoparticles are composed vs. synthesis and final applications (3). Another example is separation based on the number of deprotonated carboxylicacid groups along the backbone chain of polyacrylates and polyurethanes by applying non-aqueous ion-exchange chromatography (NAIEC). The use of the polar aprotic N-methyl-2-pyrrolidone was found to be a suitable solvent and NAIEC eluent. The retention in NAIEC was found to be guided by the number of acid groups incorporated in the copolymers. The NAIEC separation allowed the determination of the acid‑functionality distribution (AFD) of the copolymers, while incorporation of NAIEC in a 2D-LC system resulted in a fully orthogonal acid‑functionality distribution as a function of a second‑dimension MWD determination (see Figure 1a) (4). The result—the interrelated distribution of AFD with MWD (AFD×MWD)—yielded more understanding of how the incorporation of acid-functional side groups could be optimized in waterborne binder systems to balance between colloidal stabilization of the nanoparticles and optimized water sensitivity in the final application. The same hyphenated approach was applied for copolyesters, where the correlation of end-group functionality and molar mass with the chemical composition distribution was determined by the online comprehensive coupling of the mono-distribution separation for end‑group based on normal‑phase LC with SEC. The parallel ultraviolet and high‑resolution mass spectrometry detection (UV–HRMS) allowed identification and quantification of chemical composition over the combined FTD×MWD distributions (see Figure 1b) (5). The interrelated distributions could not be obtained with other analytical approaches, which rendered the developed platforms a collectively valuable tool for the further development of complex industrial copolymers.
Figure 1: (a) NAIEC×SEC of a mixture of polymers of various molar mass (polystyrene, 1056, 38.1, and 1.2 kDa at 2 mg/mL each) and polyacrylates of various acid content (PMMA-MAA [94.4] and PMMA-MAA [90/10], 20 mg/mL and 50 mg/mL, respectively) (Adapted and reproduced with permission from reference 4). (b) Normal-phase LC×SEC-UV–HRMS chromatogram of linear polyester composed of propylene glycol (PG)–terephthalic acid (TPA) copolyesters, which was derivatized with hexahydrophthalic anhydride (HHPA).
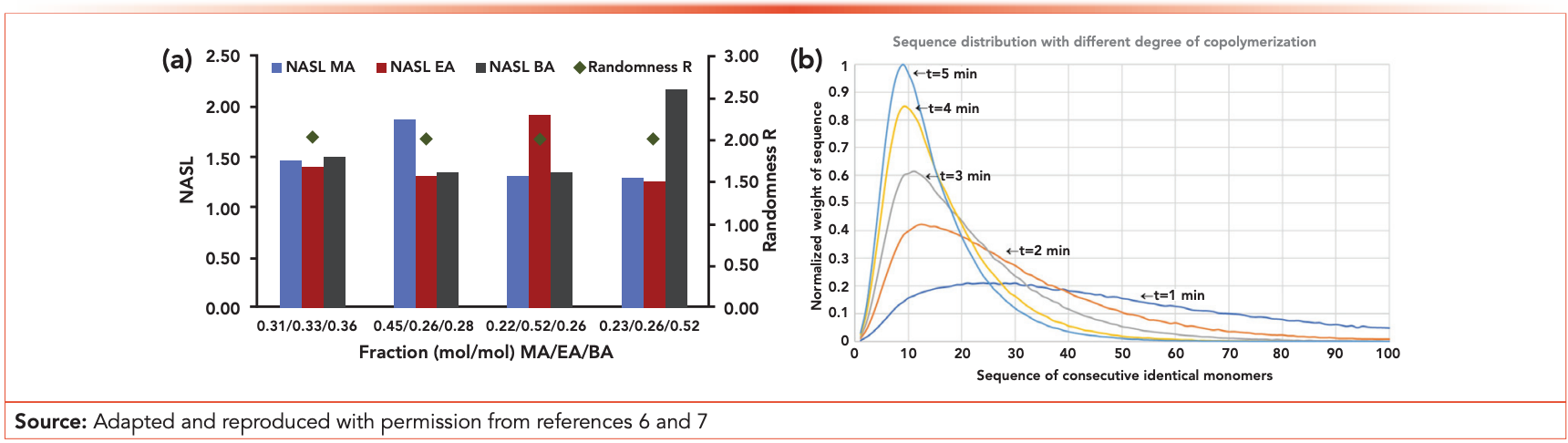
Recent developments and ongoing research towards the coupling of different information sources with respect to chemical structure distributions of copolymers have been outlined above. However, the determination of the aver‑ age block length of copolymers was a topic that received much less atten‑ tion in the past decade. While several precision synthesis approaches have become available to design copolymers with a desired sequence dis‑ tribution, an analysis of the average sequence length is challenging. As it is hard to develop separations based on the average block length of copo‑ lymers, and nuclear magnetic resonance (NMR)—the golden standard—is limited to copolymers composed of a few monomers only, further research was conducted into new approaches. In this light, a new strategy was intro‑ duced that is based on degradation of the copolymer—before or after LC separation—into short chains that contain the block length information. One example of this effort is the online coupling of LC with pyrolysis (Py). Online LC-Py analysis enables the block length determination over the MWD and chemical composition distribution of polyacrylates, for example (see Figure 2a) (6). Another example is specific MS fragmentation following LC separation. The new LC–MS fragmentation approach resulted in the block length distribution of polycondensates. Moreover, the construction of a chemo‑ metric workflow allowed the average sequence length to be converted into a sequence distribution (see Figure 2b) (7). We expect many different and interesting developments to emerge in the years to come. The result will be generalized approaches applicable for a broad set of complex industrial copolymers that will facilitate further innovation of functional and sustainable materials for use in society.
Figure 2: (a) Number average sequence length (NASL) for four tertiary polyacrylates systems based on MA, EA, and BA, as determined by online SEC-Py-MS analysis. (b) Sequence distribution as a function of the copolymerization time, as determined by LC–MS.
References
(1) Pirok, B. W. J.; Mengerink, Y.; Peters, R. A. H. Molecular Correlative Material Characterization: Advantages for Polymer Analysis Using Liquid Chromatography. LCGC Eur. 2021, 34 (5), 172–180. DOI: 10.56530/lcgc.eu.ez6278j8
(2) Pirok, B. W. J.; Abdulhussain, N.; Aalbers, T.; et al. Nanoparticle Analysis by Online Comprehensive Two-Dimensional Liquid Chromatography Combining Hydrodynamic Chromatography and Size-Exclusion Chromatography with Intermediate Sample Transformation. Anal. Chem. 2017, 89 (17), 9167–91745. DOI: 10.1021/acs.analchem.7b01906
(3) Pirok, B. W. J.; Abdulhussain, N.; Brooijmans, T.; et al. Analysis of Charged Acrylic Particles by Online Comprehensive Two-Dimensional Liquid Chromatography and Automated Data-Processing. Anal. Chim. Acta. 2019, 1054, 184–192. DOI: 10.1016/j.aca.2018.12.059
(4) Brooijmans, T.; Gonzalez, P. C.; Pirok, B. W. J.; Schoenmakers, P. J.; Peters, R. A. H. Two-Dimensional Tools for Analyzing Polymer Microstructure; Coupling Non-Aqueous Ion-Exchange Chromatography to Size-Exclusion Chromatography. J. Chromatogr. A 2022, 1683, 463536. DOI: 10.1016/j.chroma.2022.463536
(5) Molenaar, S. R. A.; van de Put, B.; Desport, J. S.; et al. Automated Feature Mining for Two-Dimensional Liquid Chromatography Applied to Polymers Enabled by Mass Remainder Analysis. Anal. Chem. 2022, 94, 14, 5599–5607. DOI: 10.1021/acs.analchem.1c05336
(6) Knol, W. C.; Vos, S.; Gruendling, T.; Pirok, B. W. J.; Peters, R. A. H. Expansion of the Application Range of Pyrolysis-Gas Chromatography to Copolymer Sequence Determination: Acrylate Copolymers. J. Anal. Appl. Pyrolysis 2022, 165, 105578. DOI: 10.1016/j.jaap.2022.105578
(7) Mengerink, Y.; Philipsen, H.; Jordens, J.; et al. Sequence Distribution Determination by SWAMP-MS a Systematic Way of Analyzing Multiple Fragmented Polymers with Mass Spectrometry. Appl. Polym. Sci. 2023, 140 (13), e53683 (2010). DOI: 10.1002/app.53683
About the Author
Ron Peters is a science fellow at Covestro. He leads the global competence development of testing, analysis, and physics in Covestro. He is also a part-time professor of bioterials analysis at the Faculty of Science (FNWI) of the University of Amsterdam (UvA), Netherlands. His current research activities focus on develop‑ ing and applying novel strategies using cutting-edge chromatographic technologies—including sample preparation, multi-separation, and detection methods—to understand the molecular structure of complex functional copolymers. Direct correspondence to: Ron.Peters@covestro.com

University of Tasmania Researchers Explore Haloacetic Acid Determiniation in Water with capLC–MS
April 29th 2025Haloacetic acid detection has become important when analyzing drinking and swimming pool water. University of Tasmania researchers have begun applying capillary liquid chromatography as a means of detecting these substances.
Prioritizing Non-Target Screening in LC–HRMS Environmental Sample Analysis
April 28th 2025When analyzing samples using liquid chromatography–high-resolution mass spectrometry, there are various ways the processes can be improved. Researchers created new methods for prioritizing these strategies.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)














