Detecting Biomarkers in Breath using GC–MS
Biomarker studies using exhaled breath are rapidly emerging as a technique for early disease detection and precision medicine. By offering a completely non-invasive experience for patients as an alternative to painful biopsy procedures. A new approach has the potential to enhance patient compliance, while making clinical workflows simpler. Exhaled breath analysis, however, requires a highly sensitive analytical technique capable of accurately measuring the broad range of volatiles present in breath. In this article, we present a proof-of-concept study to demonstrate a reliable and sensitive method to detect analytes in breath samples. Using high‑resolution accurate mass (HRAM) mass spectrometry (MS), the method validates how low- and high-abundance biomarkers can be quantified from exhaled breath.
Alila Medical Media/stock.adobe.com
A modern diagnostic technique that involves the analysis of exhaled volatile organic compounds (VOCs), which are products of metabolic activity, has been developed. In certain diseases, such as cancer, where metabolic changes can precede genetic changes, the VOCs in exhaled breath can serve as promising biomarkers for early disease detection.
Traditional biopsy methods involve removing a small amount of tissue using invasive surgical procedures, often requiring anesthesia, a team of healthcare professionals and a post-surgery recovery period. Breath analysis, on the other hand, only requires the collection of breath samples, making it completely non-invasive and pain-free for patients.
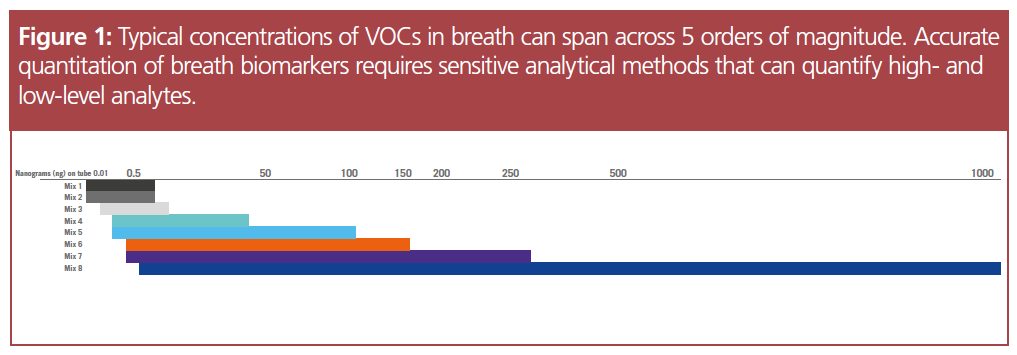
Rapidly emerging as a technique in metabolomics studies, the analysis of exhaled breath confronts a very specific analytical challenge: developing a robust method to measure the broad concentration range of different VOCs. With typical concentrations of chemicals in exhaled breath spanning five orders of magnitude, this type of analysis requires methods that can quantify both high‑abundance chemicals, as well as trace‑level VOCs at the same time.
Analytical Requirements for Breath Analysis
The analytical pre-requisite for biomarker discovery in exhaled breath involves striking the right balance between three attributes: scanning speed, selectivity (that is, high resolution and accurate mass) and sensitivity. Moreover, the diverse nature of analytes being measured also requires the method to truly represent the comprehensive range of analytes in the samples.
In the past, breath analysis employed time-of-flight (TOF) analyzers. The parallel detection of different ions in this method resulted in good sensitivity over a wide mass range, while the high resolution allowed for accurate compound identification. However, ion saturation in TOF analyzers can sometimes result in inaccurate quantitation of analytes present at higher concentrations, posing a limit on VOC analysis.
Using HRAM analyzers can address this issue because of their characteristic dynamic range. Spanning over five orders of magnitude, HRAM mass spectrometers offer a high resolving power for high- and low-concentration analytes. Additionally, the ability to detect minute differences in mass yields a sub-ppm mass accuracy, making HRAM a good method of choice to quantify trace-level chemicals in breath analysis.
Breath Analysis Workflow
A workflow for non-invasive analysis of breath biomarkers to provide early detection of disease biomarkers in conditions such as cancer, asthma, liver disease and pulmonary hypertension has been developed (1). This workflow involves collecting breath samples from patients and analyzing them using targeted or untargeted methods. In the targeted approach, samples are qualitatively and quantitatively analyzed for a list of known biomarkers, while the untargeted approach involves discovery of unknown biomarkers attributed to a disease. HRAM analyzers are particularly effective for untargeted analysis where the composition and concentration range of analytes are unknown.
A recent study provided a proof-of-concept workflow of breath sample analysis (1). With the objective of developing a reliable, sensitive and selective method to analyze unknown breath VOCs, the researchersl performed untargeted breath analysis by combining thermal desorption–gas chromatography (TD–GC) with HRAM and validated the method by testing breath samples for smoking-related markers.
Experimental Methods
Breath collection: Breath samples of 1500 mL were collected from twelve individuals, using an ReCIVA Breath Sampler (Owlstone Medical). Collected breath samples were captured and pre‑concentrated using Breath Biopsy Cartridges (Owlstone Medical) before analysis.
Sample preparation, desorption and MS analysis: Samples were dry-purged to remove excess water, desorbed using the TD100-xr automated thermal desorption system (Markes International) and transferred onto a 30 m × 0.32 mm, 3-μm column (Quadrex) using splitless injection to aid trace analysis. GC separation was achieved using a programmed method on the TRACE 1310 GC oven (Thermo Scientific) prior to MS. Internal standards used for quality control consisted of a 40-compound mixture prepared in methanol with 1 ppm, 100 ppm, and 200 ppm median concentrations. Mass spectral data was acquired on the Scientific Exactive GC Orbitrap MS (Thermo Scientific) using both variable electron ionization (EI) and chemical ionization (CI) capabilities.
Data processing: Data was acquired and processed using Xcalibur and TraceFinder software, both Thermo Fisher Scientific. Automated peak deconvolution features on TraceFinder generated clean mass spectra, while library search capabilities using custom and commercially available spectral libraries identified known and unknown compounds, giving both qualitative and quantitative outputs.
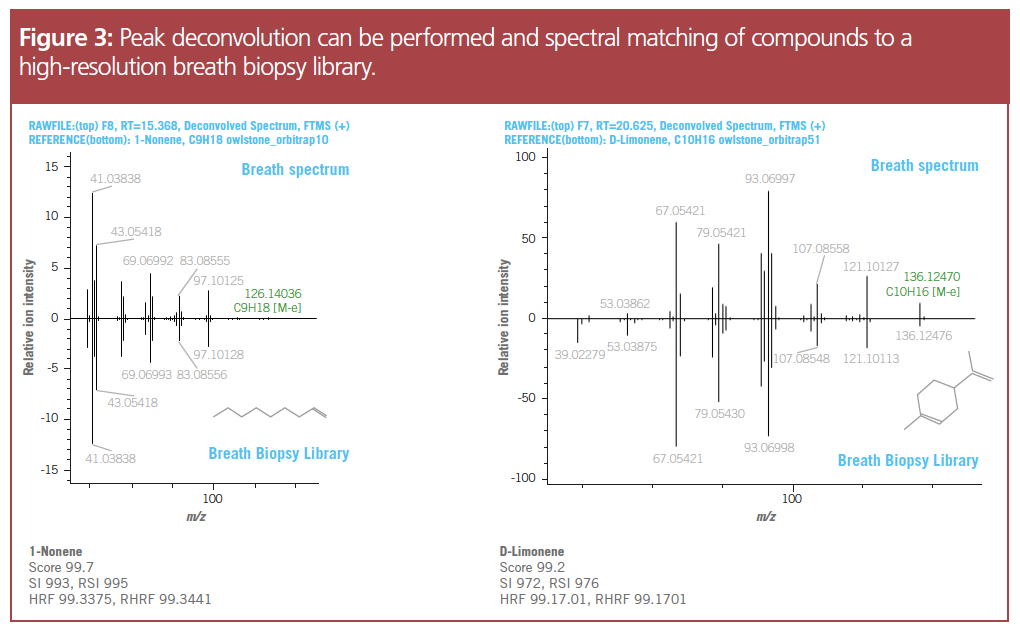
Results
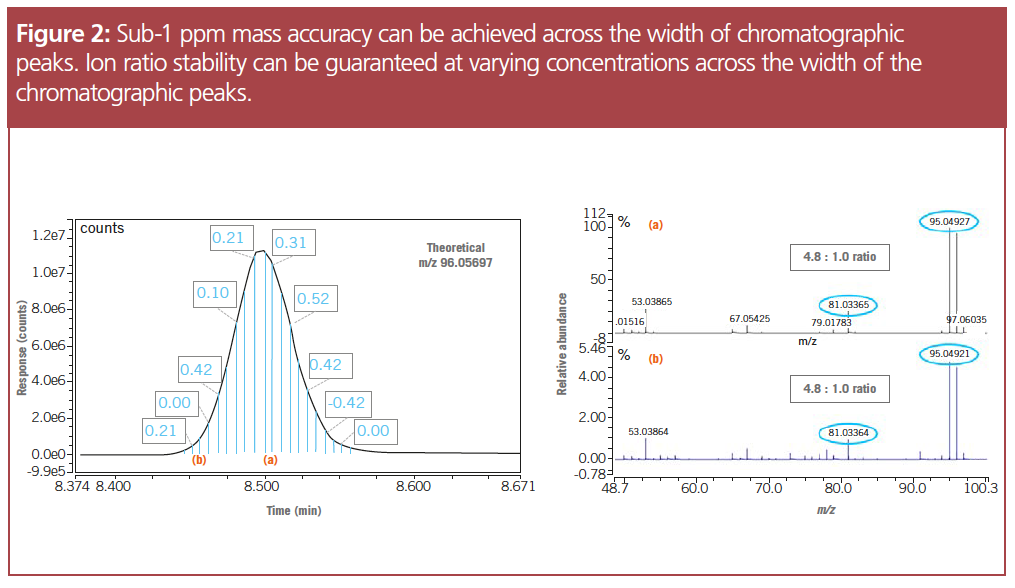
Detecting low- and high-concentration compounds in a single analysis: Sub-1 ppm mass accuracy was achieved over the full chromatographic peak, verifying the detection of low-concentration analytes using this method. The lower mass accuracy improved peak deconvolution, helping to differentiate the analytes of interest in the mixture. Identical relative fragment ion ratios were observed at both low and high concentrations. The high linear dynamic range of HRAM guarantees stable ion ratios across five orders of magnitude, again improving deconvolution and compound identification.
Precise compound identification: To enable precise identification of compounds, the software uses various data filtering scores. Peak deconvolution and spectral matching resulted in more than 1000 entries, which may be “true” peaks associated with the chemicals in breath or false positives. To accurately identify compounds in the breath samples, a mix of hard and soft ionization techniques were used for MS. High fragmentation ionization techniques, such as EI, yielded extensive fragmentation to identify closely related unknown analytes, but with a lower mass range of interpretable data. Soft ionization using CI, generated fragments of higher m/z to help differentiate between compounds within a specific class, such as alkanes or terpenes,which further improves compound identification.
Proof-of-concept: Identifying markers of smoking behaviour: The established workflow and settings on the TD–GC–HRAM system were then tested for smoking-related markers using breath samples from smokers and non-smokers as a proof-of-concept demonstration. Collected samples were divided into three groups based on smoking behaviour: never smoked (n = 3), current smokers (n = 4) and ex‑smokers (n = 5). A targeted approach was used to detect and quantify smoking‑related markers using a customized six‑compound library from previously reported data (2).
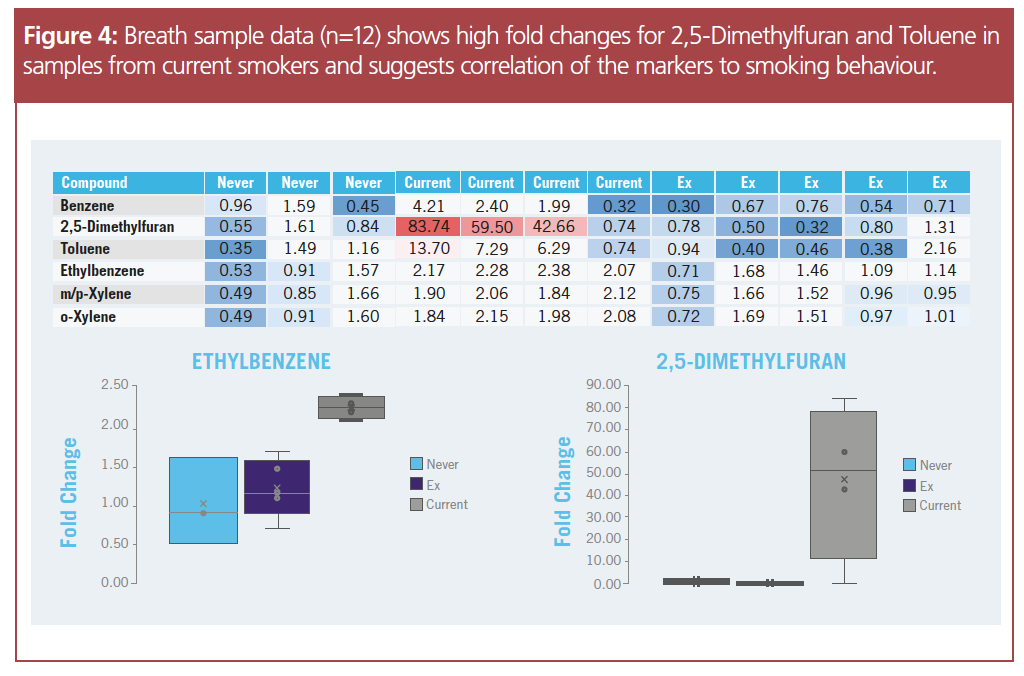
The smoking‑related markers in the database comprised of benzene, 2,5-dimethylfuran, toluene, ethylbenzene, m/p-xylene and o-xylene.
Using the above ionization and data filtering methods, fold changes for different smoking markers between the smoking and non-smoking breath samples were obtained. Low fold changes were observed in most of the smoking-related markers across all three groups, suggesting a low correlation between smoking behaviour and these reported markers. Conversely, high fold changes were observed in “current smokers” for certain markers such as 2,5-dimethylfuran and toluene, indicating a high correlation with smoking behaviour. The fold changes in smoking markers corresponding with the respective smoking behaviour demonstrates the robustness of the breath biopsy method, further corroborating that, with careful experimental design, breath volatiles can offer insights into underlying health conditions.
Conclusion
Given the need for both qualitative and quantitative data from breath samples, the TD–GC with HRAM detection met the three analytical pre-requisites for breath analysis, namely improved sensitivity, greater selectivity, and fast scanning. The broad dynamic range of five orders of magnitude and sub-ppm level sensitivity in the HRAM analyzer enabled detection of both high‑abundance and trace-level analytes in a single analysis. Features such as high mass accuracy and strong resolving power, provided improved peak deconvolution, helping to identify and quantify analytes of even trace amounts.
Data filtering algorithms, namely HRF and retention time alignment, contributed to the generation of high-quality datasets by further fine-tuning compound identification. Employing two different ionization techniques, such as EI and CI, each bringing its own area of strength, made it easier to differentiate and gain insights into the chemical structure of closely related compounds.
With untargeted discovery built into its workflow, HRAM acquires and stores the complete profile. This gives researchers the flexibility to return to the dataset at a later stage to re-analyze or even ask a different research question, allowing replication and accomplishment of longitudinal studies. When combined with phenotypic information, these virtual patient profiles can be used to test new hypothesis and augment clinical trials.
This study also demonstrates how the this workflow eliminates time-consuming sample collection and preparation steps involved in conventional, invasive biopsies, thereby saving valuable time in clinical diagnostics, and offering patients a painless experience with precision medicine through non-invasive biomarker discovery using exhaled breath.
References
- J. Boschmans, C. Cojocariu, P. Silcock, B. Boyle, A. Makarov, and M. Allsworth, “Breath biopsy: Combining thermal desorption-gas chromatography with high resolution mass spectrometry for improved sensitivity and selectivity in untargeted breath analysis”, Owlstone Medical (2018).
- S. Capone, M. Tufariello, A. Forleo, V. Longo, L. Giampetruzzi, A.V. Radogna, F. Casino, and P. Siciliano, Biomed. Chromatogr. 32(4), doi: 10.1002/bmc.4132 (2018).
Max Allsworth is the Chief Science Officer at Owlstone Medical and has over 15 years’ experience in the development of trace chemical detection technologies. He has developed sensor technologies for a vast range of applications including personal safety, food contamination and environmental monitoring.
E-mail: breathbiopsy@owlstone.co.uk
Website: www.owlstonemedical.com

University of Tasmania Researchers Explore Haloacetic Acid Determiniation in Water with capLC–MS
April 29th 2025Haloacetic acid detection has become important when analyzing drinking and swimming pool water. University of Tasmania researchers have begun applying capillary liquid chromatography as a means of detecting these substances.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)