Detailed Glycosylation Analysis of Therapeutic Enzymes Using Comprehensive 2D-LC–MS
The use of comprehensive two-dimensional liquid chromatography (LC×LC) coupled to mass spectrometry (MS) for characterizing glycosylation of therapeutic enzymes is presented. Recombinant human acid α-glucosidase (rhGAA) was digested and resulting peptides were separated by reversed-phase LC (RPLC) at high and low pH in, respectively, the first and second dimension. Glycopeptide peaks were then selectively detected and identified by MS operated in all-ion fragmentation mode. The study of first generation rhGAA (myozyme), expressed in Chinese hamster ovary (CHO) cells, and next-generation glyco-engineered rhGAA, produced in yeast cells to fine-tune the mannose-6-phosphate (M6P) content, is described.
Human acid α-glucosidase (hGAA) catalyzes the hydrolysis of glycogen to glucose in the lysosomes of the cell. There are around 50,000 people worldwide which have a deficiency of this enzyme, leading to glycogen accumulation in the lysosomes, a rare and fatal disorder known as Pompe disease (1–5). Pompe patients typically receive an enzyme replacement therapy (ERT) with recombinant human acid α-glucosidase (rhGAA) commercially known as myozyme or lumizyme. rhGAA is a heavily N-glycosylated protein with a MW of 110 kDa as expressed in Chinese hamster ovary (CHO) cells. The enzyme contains seven N-glycosylation sites which are occupied with complex and high mannose glycans (2–5). The former complex glycans are predominantly sialylated and, to a lesser extent, acetylated, the latter glycans contain mannose-6-phosphate (M6P) structures considered a critical quality attribute (CQA) as these are responsible for targeting the enzyme to the lysosomal compartment of the cell where it needs to be catalytically active and break down glycogen.
To study N-glycosylation of therapeutic enzymes, glycans are commonly liberated from the protein backbone using PNGase F, fluorescently labeled and separated using hydrophilic interaction liquid chromatography (HILIC) (6–9). While this methodology provides a wealth of information, site specific data is lost, such as which glycans are conjugated to which asparagine residue, and to which extent. To obtain the latter, peptide mapping is required. When digesting a therapeutic enzyme, hundreds of peptides are to be expected with varying physicochemical properties present in a wide concentration range. Two-dimensional liquid chromatography (2D-LC) is perfectly suited to tackle this complexity (10). In 2D-LC, two different chromatographic separation mechanisms are combined and material eluted from a first column is further separated on a second column which has an orthogonal separation behavior. 2D-LC comes in different flavors: (multiple-)heart-cutting (LC−LC), where one or a couple of peaks are transferred from first to second dimension, and comprehensive 2D-LC (LC×LC) where the entire first dimension chromatogram is sampled thereby maximizing separation power. The present manuscript demonstrates how LC×LC in combination with quadrupole-time-of-flight (Q-TOF) mass spectrometry (MS) operated in all-ion fragmentation mode comes in as a very powerful tool to study glycosylation of rhGAA.
Materials and Methods
Materials
Water, acetonitrile (ACN), methanol (MeOH) and formic acid were purchased from Biosolve. Dithiothreitol (DTT), 2-iodoacetamide (IAA) and ammonium bicarbonate were from Sigma-Aldrich. Tris-HCl pH 8 was purchased as a 1M solution from Thermo Fisher Scientific. Porcine sequencing grade modified trypsin was acquired from Promega and Rapigest from Waters. Myozyme was obtained from Sanofi-Genzyme. rhGAA was expressed in glyco-engineered Yarrowia lipolytica, essentially as described in Tiels et al (11).
Sample Preparation
To a volume corresponding to 100 µg of therapeutic enzyme, 105 µL of 0.1% Rapigest in 100 mM Tris-HCl pH 8 was added followed by the addition of 100 mM Tris-HCl pH 8 to a final volume of 192.5 µL. The sample was subsequently reduced at 60 °C for 30 min by the addition of 5 mM DTT (2.5 µL of 400 mM DTT in 100 mM Tris-HCl pH 8) and alkylated at 37 °C for 1 h by adding 10 mM IAA (5 µL of 400 mM IAA in 100 mM Tris-HCl pH 8). Lyophilized trypsin (20 µg) dissolved in 100 mM Tris-HCl pH 8 (50 µL) was added in a volume of 10 µL, giving rise to a final sample volume of 210 µL and an enzyme to substrate ratio of 1/25 (w/w). Digestion proceeded for 16 h at 37 °C.
LC×LC–MS
LC×LC analyses were carried out on an Agilent 1290 Infinity 2D-LC system (Agilent Technologies). Two G4220A binary pumps, a G4226A autosampler with G1330A autosampler thermostat, a G1316C thermostatted column compartment and a G1170A valve drive with 2-position/4-port duo valve (G4236A), equipped with two 40 µL loops, were used. High-resolution accurate mass data were acquired on an Agilent G6530 Q-TOF equipped with a JetStream source (Agilent Technologies). The 2D-LC system was controlled by OpenLab CDS Chemstation with 2D-LC add-on software and the Q-TOF by MassHunter Acquisition software (Agilent Technologies). Data analysis was performed with GC Image LC×LC-HRMS Edition software (GC Image, LLC) and MassHunter Qualitative Analysis software complemented with BioConfirm (Agilent Technologies). LC×LC and MS method details are listed in Table I. To cope with the high flow rate, the effluent of the second dimension column was split. Therefore, a zero dead volume T-piece was connected to the second dimension column outlet via a stainless steel capillary (9 mm long, 0.12 mm internal diameter). The outlets of the T-piece were connected to the MS (directly on the ESI needle) with stainless steel tubing (34 cm long, 0.075 mm internal diameter) and to the waste with a stainless steel capillary (27 cm long, 0.12 mm internal diameter). In this manner, the necessary restriction is provided to direct the majority of the flow to the waste and only a small fraction to the MS.

Results and Discussion
In this study, site-specific glycosylation of rhGAA was studied using LC×LC. Compared to one-dimensional LC (1D-LC), resolution in LC×LC is drastically increased as long as the two dimensions are orthogonal and separation obtained in the first dimension is maintained upon transfer to the second dimension. To achieve the latter, effluent is collected in two loops installed on a 2-position valve which are alternately transferred to the second dimension column. The second dimension analysis of one loop takes place during the filling of the other loop. As such, there is a demand for fast second dimension separations to maintain the first dimension separation. The selectivity of the two separation mechanisms toward the peptides must differ substantially in order to maximize orthogonality and resolution. The combination of reversed-phase LC (RPLC) at pH extremes—high pH in first and low pH in second dimension, is very powerful in that respect (12–14). Orthogonality is mainly directed by the mobile phase pH and the zwitterionic nature of the peptides. Partial correlation that might exist is overruled by the high peak capacity in both dimensions. Using a shifting second dimension gradient with increasing elution strength in function of analysis time furthermore increases surface coverage.
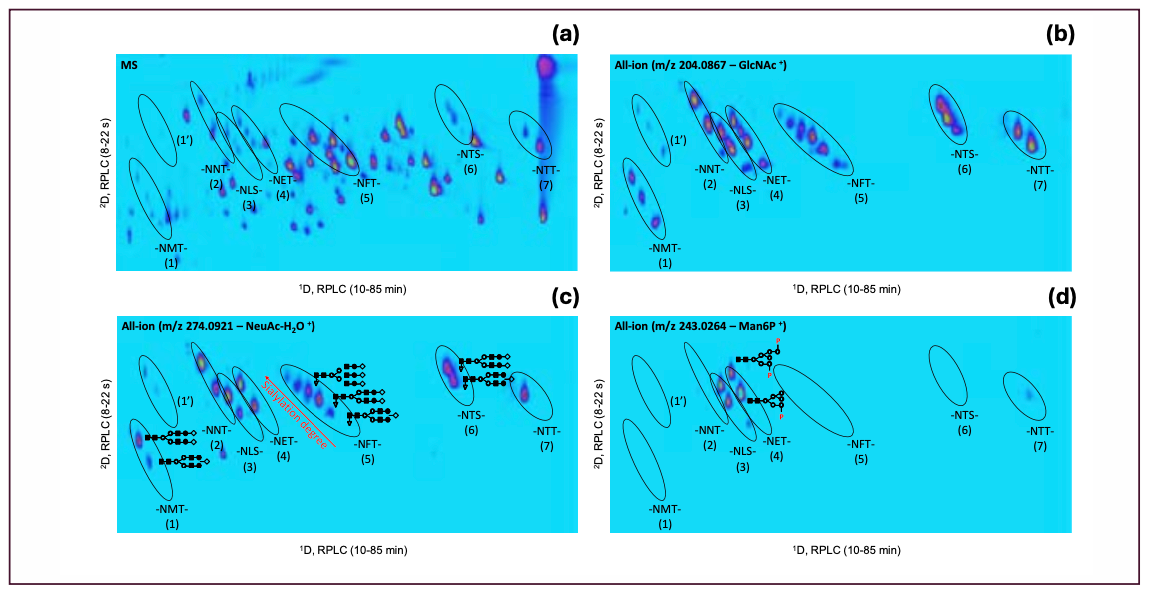
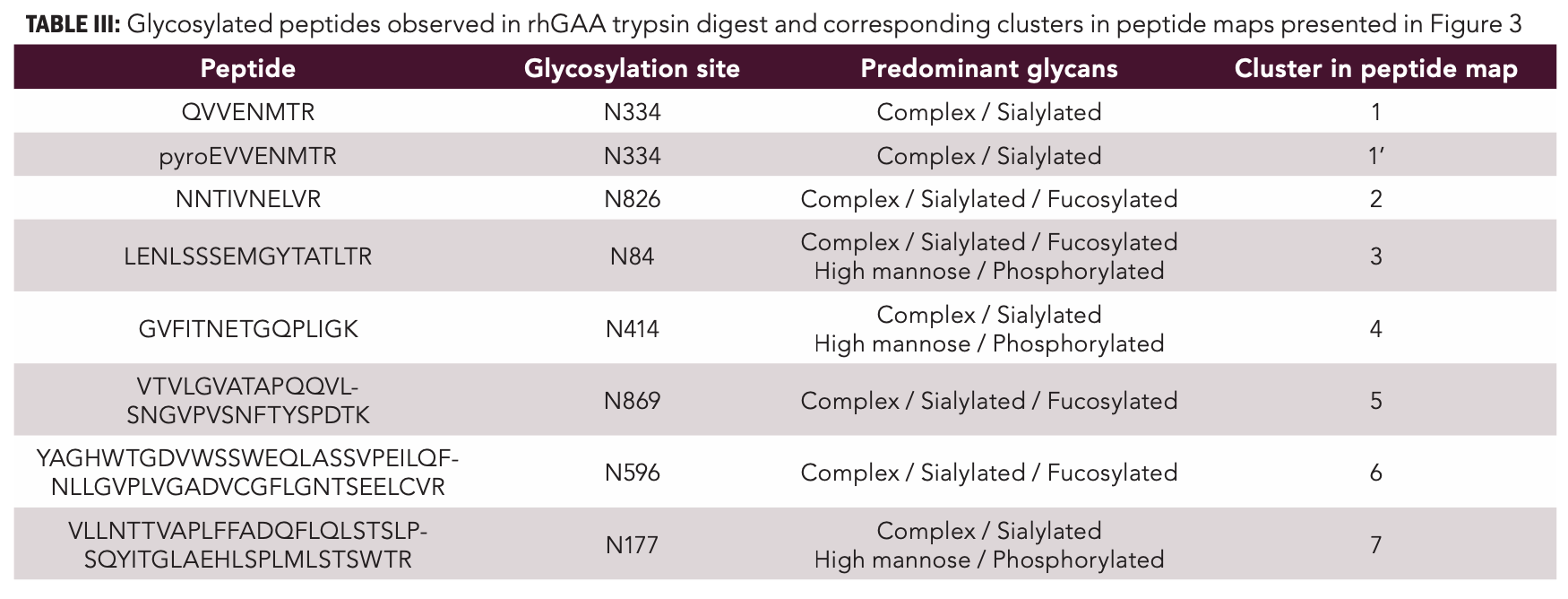
Figure 1 shows the RPLC×RPLC–MS peptide map of the myozyme digest. Excellent chromatographic and mass spectrometric performance is obtained providing detailed structural insights. The MS/MS spectra of two representative N-glycosylated peptides are shown in Figure 2. Upon collision induced dissociation (CID), glycosylated peptides give rise to specific fragments originating from the glycan part. These sugar oxonium ions (Table II) can be used to selectively recognize glycosylated peptides in the data. For that, one can operate the mass spectrometer in the all-ion fragmentation mode in which all peptides are transferred into the CID cell where they are fragmented. By alternating all-ion fragmentation with regular MS acquisition, precursors giving rise to sugar oxonium ions can be revealed and a detailed study of glycosylation sites achieved. Figures 3a-d show the regular MS peptide map (a) and the all-ion fragmentation peptide map extracting the sugar oxonium ions at m/z 204.0867 (b), 274.0921 (c), and 243.0264 (d). The ion at m/z 204.0867 corresponding to N-acetylglucosamine is shared by all N-glycans and can be used as a general marker for N-glycosylation. Consequently, Figure 3b reveals all glycosylated peptides. Different clusters are observed in the peptide map which in fact correspond to the seven different glycosylation sites (Table III). Note that glycosylation site N334 is spread over two regions (1 and 1’) because of the partial cyclization of the N-terminal amino acid glutamine (formation of pyroglutamate–pyroE–during trypsin digestion rendering glycopeptide more hydrophobic). The different spots within a cluster correspond to different glycans at a given glycosylation site. When extracting the oxonium ions at m/z 274.0921 and 243.0264 (Figures 3 c-d), one can specifically visualize peptides decorated with, respectively, sialylated (N-acetylneuraminic acid-NeuAc) and phosphorylated (M6P) N-glycans. The data reveals that three sites (N84, N177, and N414) are occupied with phosphorylated glycans while all sites are decorated with sialylated N-glycans (Table III). A remarkable separation is achieved based on site heterogeneity with glycopeptides eluting in the following order in the first dimension at high pH: di-phosphorylated < di-sialylated < mono-phosphorylated < mono-sialylated < neutral. In the second dimension at low pH, elution order is reversed: neutral < mono-sialylated < mono-phosphorylated < di-sialylated < di-phosphorylated.
FIGURE 1: LC×LC–MS peptide map of rhGAA trypsin digest using RPLC at high pH in first (1D) and RPLC at low pH in second dimension (2D).

FIGURE 2: MS/MS spectra of tryptic peptide GVFITNETGQPLIGK decorated with phosphorylated high mannose (a) and sialylated complex N-glycans (b).
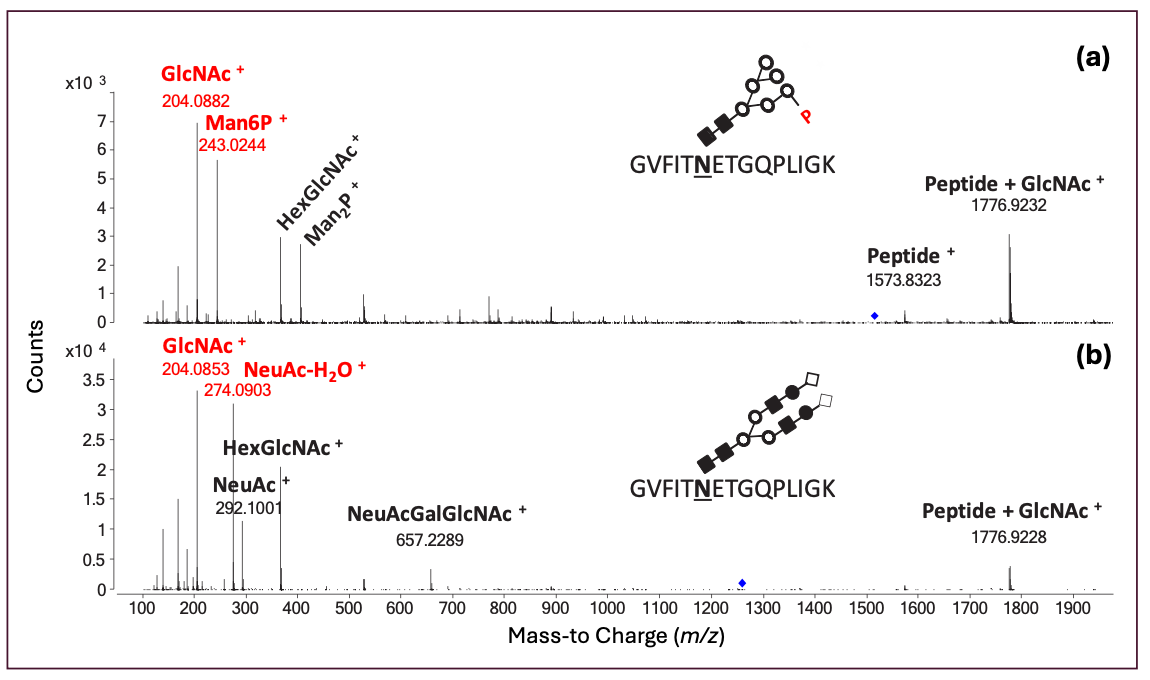
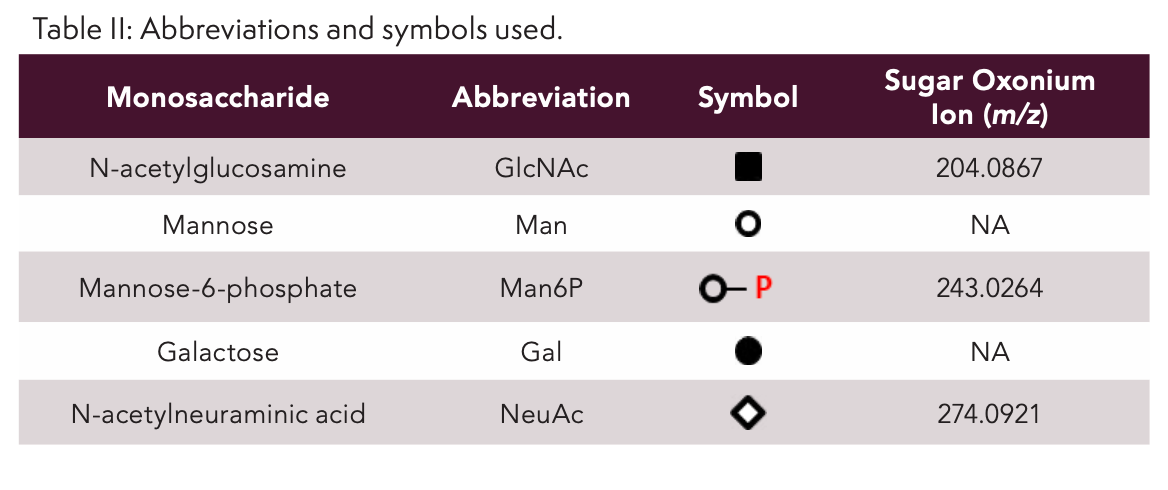
FIGURE 3: LC×LC–MS peptide map of rhGAA digest. (a) Full MS data; (b) all-ion fragmentation data extracting the sugar oxonium ions at m/z 204.0867 for N-acetylglucosamine-GlcNAc; (c) m/z 274.0921 for N-acetylneuraminic acid-NeuAc (c); and (d) m/z 243.0264 for mannose-6-phosphate–Man6P. Ions were extracted at 20 ppm mass accuracy. Clusters 1–7 correspond to specific glycosylation sites (see Table III) and different spots within a given cluster to different glycans at a given glycosylation site.
While M6P is required for targeting the enzyme to the lysosomes, only three N-glycosylation sites decorated with the latter species are present in myozyme, resulting in poor cellular uptake (11). Several next-generation glyco-engineered variants with improved cellular uptake have been developed where M6P content is increased by (1) conjugating pre-synthesized phosphorylated oligosaccharides to oxidized sialylated complex N-glycans, or (2) by expression in glyco-engineered yeast cells that have been modified to produce phosphorylated high mannose N-glycans, and subsequent exposing of the phosphate and trimming terminal mannose residues to generate the desired N-glycan structures (11,15). Figures 4a-d presents the RPLC×RPLC–MS peptide map of the glyco-engineered variant produced in yeast cells in comparison to the first-generation product derived from CHO cell expression (full MS and all-ion fragmentation). It can be concluded that yeast-derived rhGAA is devoid of sialylated N-glycans and occupied with phosphorylated high-mannose N-glycans at all seven N-glycosylation sites. In-vitro measurements (Figure 5) furthermore demonstrate that the glyco-engineered variant gives rise to a 20-fold higher uptake in fibroblasts from Pompe patients with the increased M6P content responsible for this outcome. This represents a perfect illustration of how cutting-edge biology and state-of-the-art analytics go hand in hand.
FIGURE 4: LC×LC–MS peptide map of (a,c,e,g) first- and (b,d,f,h) next-generation glyco-engineered rhGAA digest. (a-b) Full MS data; (c-d) all-ion fragmentation data extracting the sugar oxonium ions at m/z 204.0867 for N-acetylglucosamine-GlcNAc; (e-f) m/z 274.0921 for N-acetylneuraminic acid-NeuAc; (g-h) m/z 243.0264 for mannose-6-phosphate–Man6P. Ions were extracted at 20 ppm mass accuracy. Additional spots revealed in Figure 4h can be traced back to M6P carrying O-glycosylated peptides.

FIGURE 5: Enzyme uptake in Pompe disease patient fibroblasts. Experimental details can be found in Tiels et al. (11).

Conclusion
The detailed study of glycosylation of the therapeutic enzyme rhGAA using LC×LC–MS has been demonstrated. The resolving power offered facilitates an in-depth structural characterization and using all-ion fragmentation, glycosylated peptides can selectively be recognized facilitating data interpretation. The method has successfully been applied to first and next-generation rhGAA and findings placed in a biological context.
References
(1) Roig-Zamboni, V.; Cobucci-Ponzano, B.; Iacono, R.; Ferrara, M.C., Germany, S.; Bourne, Y.; Parenti, G.; Moracci, M.; Sulzenbacher, G. Structure of Human Lysosomal Acid α-glucosidase-A Guide for the Treatment of Pompe Disease. Nat. Commun. 2017, 8, 1111. DOI: 10.1038/s41467-017-01263-3
(2) McVie-Wylie, A.J.; Lee, K.L.; Qiu, H.; Jin, X.; Do, H.; Gotschall, R.; Thurberg, B.L.; Rogers, C.; Raben, N.; O’Callaghan, M.; Canfield, W.; Andrews, L.; McPherson, J.M.; Mattaliano, R.J. Biochemical and Pharmacological Characterization of Different Recombinant Acid Alpha-Glucosidase Preparations Evaluated for the Treatment of Pompe Disease. Mol. Genet. Metab. 2008, 94, 448–455. DOI: 10.1016/j.ymgme.2008.04.009
(3) Park, H.; Kim, J.. Lee, Y.K.; Kim, W.; You, S.K.; Do, J.; Jang, Y.; Oh D.B.; Il Kim, J.; Kim, H.H. Four Unreported Types of Glycans Containing Mannose-6-phosphate are Heterogeneously Attached at Three Sites (Including Newly Found Asn 233) to Recombinant Human Acid Alpha-Glucosidase That is the Only Approved Treatment for Pompe Disease. Biochem. Biophys. Res. Commun. 2018, 495, 2418–2424. DOI: 10.1016/j.bbrc.2017.12.101
(4) Park, H.; You, S.; Kim, J.; Kim, W.; Do, J.; Jang, Y.; Kim, D.; Lee, J.; Ha, J.; Oh, D.B.; Kim, J.I.; Kim, H.H. Seventeen O-acetylated N-glycans and Six O-acetylation sites of Myozyme Identified Using Liquid Chromatography-Tandem Mass Spectrometry. J. Pharm. Biomed. Anal. 2019, 169, 188–195. DOI: 10.1016/j.jpba.2019.03.013
(5) Di Marco, F.; Blöchl, C.; Esser-Skala, W.; Schäpertöns, V.; Zhang, T.; Wuhrer, M.; Sandra, K.; Wohlschlager, T.; Huber, C.G. Glycoproteomics of a Single Protein: Revealing Tens of Thousands of Myozyme Glycoforms by Hybrid HPLC–MS Approaches. Mol. Cell. Proteomics 2023, 22, 100622. DOI: 10.1016/j.mcpro.2023.100622
(6) Sandra, K.; Vandenheede, I.; Sandra, P. Modern Chromatographic and Mass Spectrometric Techniques for Protein Biopharmaceutical Characterization. J. Chromatogr. A 2014, 1335, 81–103. DOI: 10.1016/j.chroma.2013.11.057
(7) Fekete, S.; Guillarme, D.; Sandra, P.; Sandra, K. Chromatographic, Electrophoretic, and Mass Spectrometric Methods for the Analytical Characterization of Protein Biopharmaceuticals. Anal. Chem. 2016, 88, 480–507. DOI: 10.1021/acs.analchem.5b04561
(8) D’Atri, V.; Dumont, E.; Vandenheede, I.; Guillarme, D.; Sandra, P.; Sandra, K. Hydrophilic Interaction Chromatography for the Characterization of Therapeutic Monoclonal Antibodies at Protein, Peptide, and Glycan Levels. LCGC Europe 2017, 30, 424.
(9) Vandenbussche, J.; Sandra, P.; Sandra, K. Analyzing Phosphorylated N-glycans with Recovery on Bio-Inert LC Systems and PEEK-lined HILIC Columns. LCGC Europe 2018, 31, 566–571.
(10) Sandra, K.; Vandenheede, I.; Steenbeke, M.; Vanhoenacker, G.; Sandra, P. Characterizing Monoclonal Antibodies and Antibody-Drug Conjugates Using 2D-LC–MS. LCGC Europe 2017, 30, 149–157.
(11) Tiels, P.; Baranova, E.; Piens, K.; De Visscher, C.; Pynaert, G.; Nerinckx, W.; Stout, J.; Fudalej, F.; Hulpiau, P.; Tännler, S.; Geysens, S.; Van Hecke, A.; Valevska, A.; Vervecken W.; Remaut, H.; Callewaert, N. A Bacterial Glycosidase Enables Mannose-6-phosphate Modification and Improved Cellular Uptake of Yeast-Produced Recombinant Human Lysosomal Enzymes. Nat. Biotechnol. 2012, 30, 1225–1231. DOI: 10.1038/nbt.2427
(12) Vanhoenacker, G.; Vandenheede, I.; David, F.; Sandra, P.; Sandra, K. Comprehensive Two-Dimensional Liquid Chromatography of Therapeutic Monoclonal Antibody Digests. Anal. Bioanal. Chem. 2015, 407, 355–366. DOI: 10.1007/s00216-014-8299-1
(13) Sandra, K.; Vanhoenacker, G.; Vandenheede, I. Steenbeke, M.; Joseph, M.; Sandra, P. Multiple Heart-Cutting and Comprehensive Two-Dimensional Liquid Chromatography Hyphenated to Mass Spectrometry for the Characterization of the Antibody-Drug Conjugate Ado-trastuzumab Emtansine. J. Chromatogr. B 2016, 1032, 119–130. DOI: 10.1016/j.jchromb.2016.04.040
(14) Sandra, K.; Steenbeke, M.; Vandenheede, I.; Vanhoenacker, G.; Sandra, P. The Versatility of Heart-Cutting and Comprehensive Two-Dimensional Liquid Chromatography in Monoclonal Antibody Clone Selection. J. Chromatogr. A 2017, 1523, 283–292. DOI: 10.1016/j.chroma.2017.06.052
(15) Oh, D.B. Glyco-Engineering Strategies for the Development of Therapeutic Enzymes with Improved Efficacy for the Treatment of Lysosomal Storage Diseases. BMB Rep. 2015, 48, 438–444. DOI: 10.5483/BMBRep.2015.48.8.101
About the Authors
Koen Sandra is the editor of “Biopharmaceutical Perspectives”. He is the CEO of RIC group (Kortrijk, Belgium) and Visiting Professor at Ghent University (Ghent, Belgium). He is also a member of LCGC International’s editorial advisory board. Kathleen Piens was Head of Analytics at Oxyrane (Ghent, Belgium) and currently holds the position of Head of Downstream Processing at Those Vegan Cowboys (Ghent, Belgium). Debby Bracke was Scientist Analytics at Oxyrane and is now Senior Scientist Discovery at Zomagen Biosciences (Ghent, Belgium). Pat Sandra is the Founder and Advisor of the RIC group and Emeritus Professor at Ghent University. Wouter Vervecken is the CEO of Oxyrane.
Direct correspondence to: koen.sandra@ric-group.com

Regulatory Deadlines and Supply Chain Challenges Take Center Stage in Nitrosamine Discussion
April 10th 2025During an LCGC International peer exchange, Aloka Srinivasan, Mayank Bhanti, and Amber Burch discussed the regulatory deadlines and supply chain challenges that come with nitrosamine analysis.
Polysorbate Quantification and Degradation Analysis via LC and Charged Aerosol Detection
April 9th 2025Scientists from ThermoFisher Scientific published a review article in the Journal of Chromatography A that provided an overview of HPLC analysis using charged aerosol detection can help with polysorbate quantification.
Removing Double-Stranded RNA Impurities Using Chromatography
April 8th 2025Researchers from Agency for Science, Technology and Research in Singapore recently published a review article exploring how chromatography can be used to remove double-stranded RNA impurities during mRNA therapeutics production.












